Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T33124
(Former ID: TTDI02364)
|
|||||
| Target Name |
D-amino acid oxidase (DAO)
|
|||||
| Synonyms |
Daminoacid oxidase; DAMOX; DAAO
Click to Show/Hide
|
|||||
| Gene Name |
DAO
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Ataxic disorder [ICD-11: 8A03] | |||||
| 2 | Schizophrenia [ICD-11: 6A20] | |||||
| Function |
Regulates the level of the neuromodulator D-serine in the brain. Has high activity towards D-DOPA and contributes to dopamine synthesis. Could act as a detoxifying agent which removes D-amino acids accumulated during aging. Acts on a variety of D-amino acids with a preference for those having small hydrophobic side chains followed by those bearing polar, aromatic, and basic groups. Does not act on acidic amino acids.
Click to Show/Hide
|
|||||
| BioChemical Class |
CH-NH(2) donor oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.4.3.3
|
|||||
| Sequence |
MRVVVIGAGVIGLSTALCIHERYHSVLQPLDIKVYADRFTPLTTTDVAAGLWQPYLSDPN
NPQEADWSQQTFDYLLSHVHSPNAENLGLFLISGYNLFHEAIPDPSWKDTVLGFRKLTPR ELDMFPDYGYGWFHTSLILEGKNYLQWLTERLTERGVKFFQRKVESFEEVAREGADVIVN CTGVWAGALQRDPLLQPGRGQIMKVDAPWMKHFILTHDPERGIYNSPYIIPGTQTVTLGG IFQLGNWSELNNIQDHNTIWEGCCRLEPTLKNARIIGERTGFRPVRPQIRLEREQLRTGP SNTEVIHNYGHGGYGLTIHWGCALEAAKLFGRILEEKKLSRMPPSHL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T36HAE | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | TAK-831 | Drug Info | Phase 2 | Friedreich's ataxia | [2] | |
| 2 | TAK-831 | Drug Info | Phase 2 | Friedreich's ataxia | [3] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | SEP-227900 | Drug Info | Terminated | Cognitive impairment | [4] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 3 Inhibitor drugs | + | ||||
| 1 | TAK-831 | Drug Info | [1], [2] | |||
| 2 | TAK-831 | Drug Info | [5] | |||
| 3 | SEP-227900 | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Flavin-Adenine Dinucleotide | Ligand Info | |||||
| Structure Description | Crystal structure of human D-amino acid oxidase in complex with inhibitor | PDB:7U9U | ||||
| Method | X-ray diffraction | Resolution | 1.66 Å | Mutation | No | [6] |
| PDB Sequence |
MRVVVIGAGV
10 IGLSTALCIH20 ERYHSVLQPL30 DIKVYADRFT40 PLTTTDVAAG50 LWQPYLSDPN 60 NPQEADWSQQ70 TFDYLLSHVH80 SPNAENLGLF90 LISGYNLFHE100 AIPDPSWKDT 110 VLGFRKLTPR120 ELDMFPDYGY130 GWFHTSLILE140 GKNYLQWLTE150 RLTERGVKFF 160 QRKVESFEEV170 AREGADVIVN180 CTGVWAGALQ190 RDPLLQPGRG200 QIMKVDAPWM 210 KHFILTHDPE220 RGIYNSPYII230 PGTQTVTLGG240 IFQLGNWSEL250 NNIQDHNTIW 260 EGCCRLEPTL270 KNARIIGERT280 GFRPVRPQIR290 LEREQLRTGP300 SNTEVIHNYG 310 HGGYGLTIHW320 GCALEAAKLF330 GRILEEKKLS340
|
|||||
|
|
ILE6
3.197
GLY7
2.104
ALA8
1.929
GLY9
2.374
VAL10
2.842
ILE11
2.222
GLY12
3.289
TYR35
3.826
ALA36
2.525
ASP37
2.086
ARG38
1.801
PHE39
3.831
THR43
2.608
THR44
1.706
THR45
1.842
ASP46
4.673
VAL47
2.265
ALA48
1.825
ALA49
3.270
GLY50
2.183
LEU51
1.970
TRP52
3.858
GLN53
3.960
LEU145
4.558
ARG162
2.440
LYS163
2.791
VAL164
1.956
GLU165
4.144
CYS181
2.912
THR182
2.419
GLY183
2.956
VAL184
4.221
TRP185
2.291
ALA188
3.505
LEU189
2.723
GLY198
4.156
ARG199
4.583
GLY200
2.314
GLN201
3.971
ILE202
2.170
TYR228
3.101
ILE230
4.227
THR237
4.571
ILE241
4.755
THR280
4.703
GLY281
2.520
PHE282
3.439
ARG283
2.500
PRO284
2.629
TYR309
4.327
GLY310
4.563
HIS311
2.744
GLY312
1.779
GLY313
2.399
TYR314
2.446
GLY315
2.192
LEU316
2.613
THR317
1.819
ILE318
4.534
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: 4H-thieno[3,2-b]pyrrole-5-carboxylic acid | Ligand Info | |||||
| Structure Description | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | PDB:3ZNN | ||||
| Method | X-ray diffraction | Resolution | 1.90 Å | Mutation | No | [7] |
| PDB Sequence |
MRVVVIGAGV
10 IGLSTALCIH20 ERYHSVLQPL30 DIKVYADRFT40 PLTTTDVAAG50 LWQPYLSDPN 60 NPQEADWSQQ70 TFDYLLSHVH80 SPNAENLGLF90 LISGYNLFHE100 AIPDPSWKDT 110 VLGFRKLTPR120 ELDMFPDYGY130 GWFHTSLILE140 GKNYLQWLTE150 RLTERGVKFF 160 QRKVESFEEV170 AREGADVIVN180 CTGVWAGALQ190 RDPLLQPGRG200 QIMKVDAPWM 210 KHFILTHDPE220 RGIYNSPYII230 PGTQTVTLGG240 IFQLGNWSEL250 NNIQDHNTIW 260 EGCCRLEPTL270 KNARIIGERT280 GFRPVRPQIR290 LEREQLRTGP300 SNTEVIHNYG 310 HGGYGLTIHW320 GCALEAAKLF330 GRILEEKKLS340
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
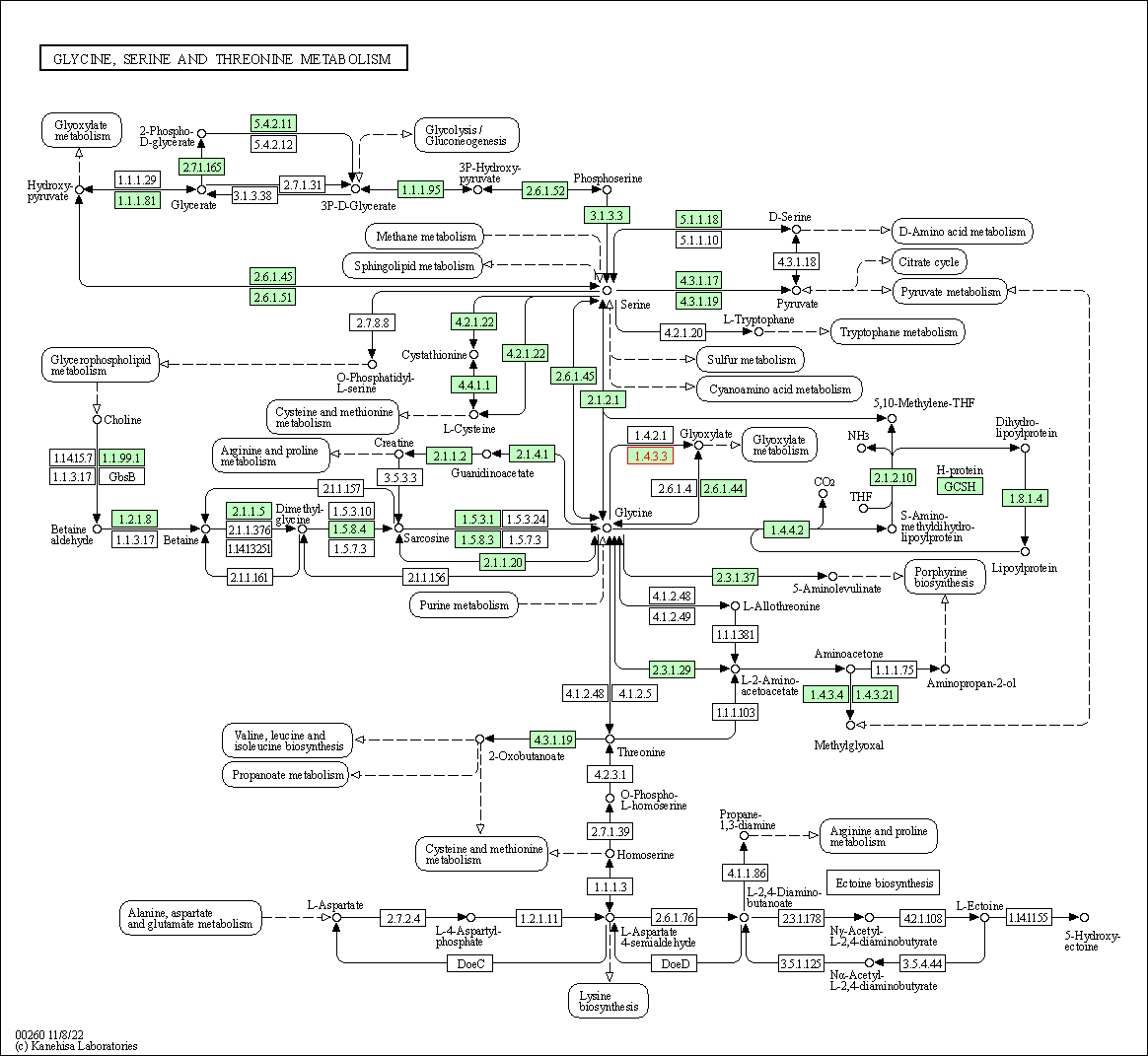
| Glycine, serine and threonine metabolism | hsa00260 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
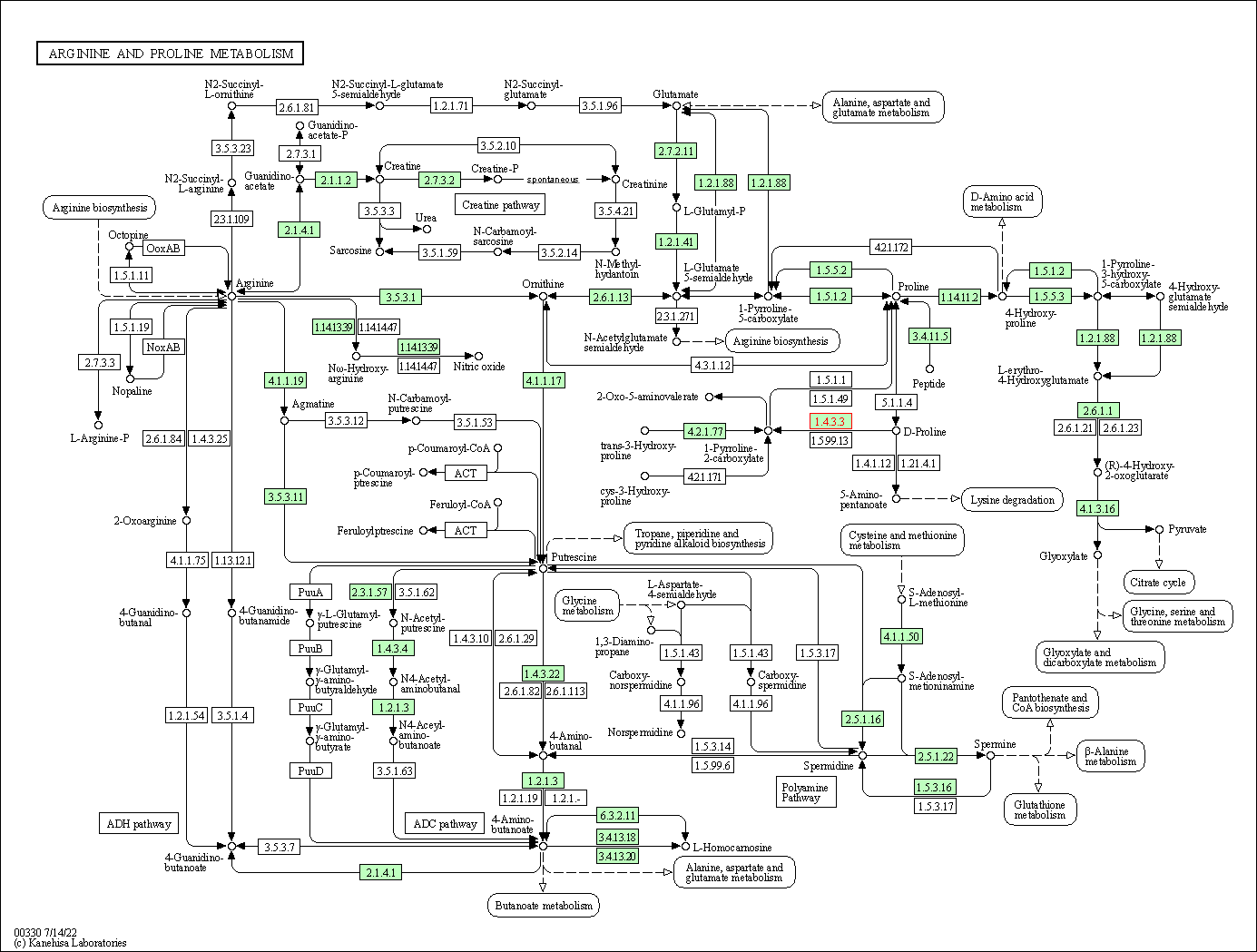
| Arginine and proline metabolism | hsa00330 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
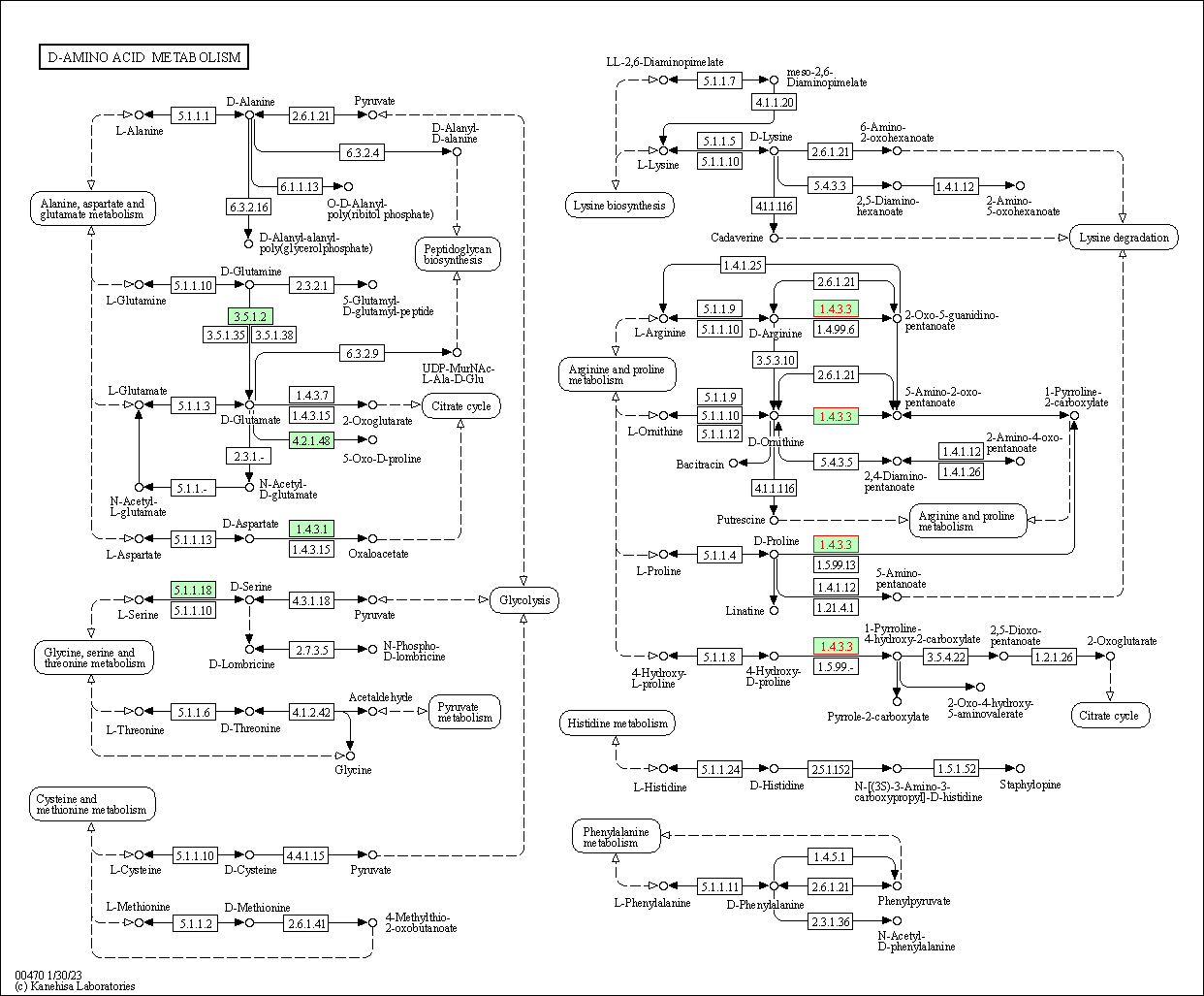
| D-Amino acid metabolism | hsa00470 | Affiliated Target |

|
| Class: Metabolism => Metabolism of other amino acids | Pathway Hierarchy | ||
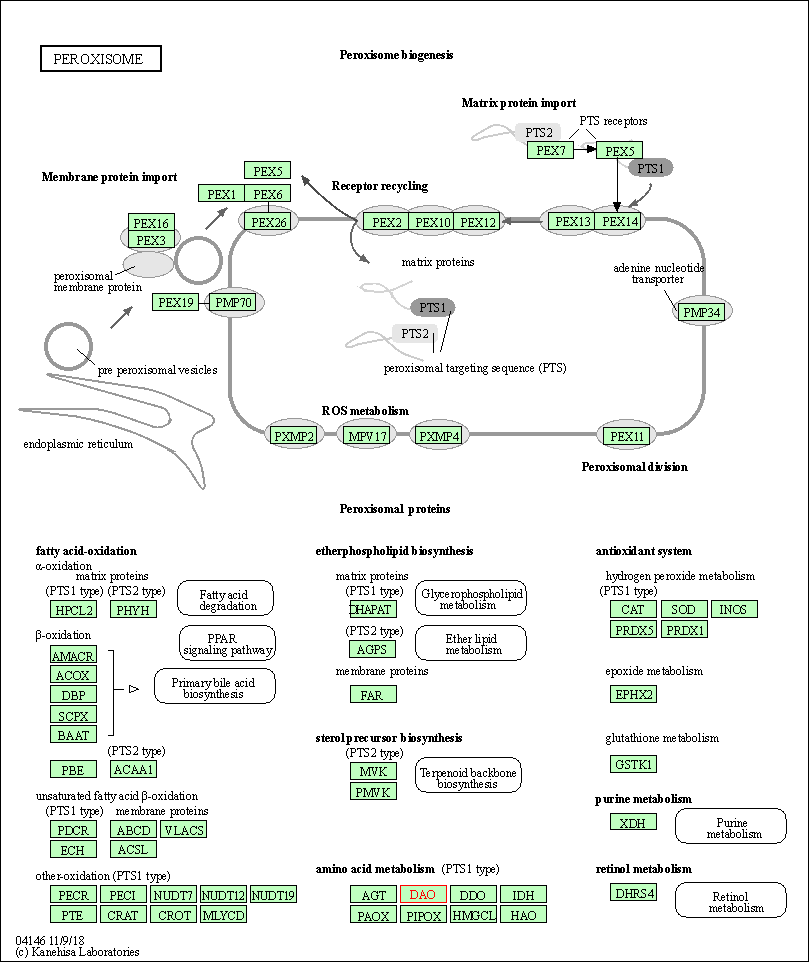
| Peroxisome | hsa04146 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Degree | 6 | Degree centrality | 6.45E-04 | Betweenness centrality | 5.16E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.91E-01 | Radiality | 1.33E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 1.48E+01 | Topological coefficient | 2.96E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 6 KEGG Pathways | + | ||||
| 1 | Glycine, serine and threonine metabolism | |||||
| 2 | Arginine and proline metabolism | |||||
| 3 | D-Arginine and D-ornithine metabolism | |||||
| 4 | Metabolic pathways | |||||
| 5 | Biosynthesis of antibiotics | |||||
| 6 | Peroxisome | |||||
| Pathwhiz Pathway | [+] 2 Pathwhiz Pathways | + | ||||
| 1 | Arginine and Proline Metabolism | |||||
| 2 | D-Arginine and D-Ornithine Metabolism | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Metabolism of amino acids and derivatives | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | ClinicalTrials.gov (NCT03214588) Efficacy, Tolerability, and Pharmacokinetics of Multiple Doses of Oral TAK-831 in Adults With Friedreich Ataxia. U.S. National Institutes of Health. | |||||
| REF 4 | The Therapeutic Potential of D-Amino Acid Oxidase (DAAO) Inhibitors. Open Med Chem J. 2010; 4: 3-9. | |||||
| REF 5 | Mechanistic Multilayer Quantitative Model for Nonlinear Pharmacokinetics, Target Occupancy and Pharmacodynamics (PK/TO/PD) Relationship of D-Amino Acid Oxidase Inhibitor, TAK-831 in Mice. Pharm Res. 2020 Aug 5;37(8):164. | |||||
| REF 6 | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schr?dinger Computational Platform. J Med Chem. 2022 May 12;65(9):6775-6802. | |||||
| REF 7 | Structural, kinetic, and pharmacodynamic mechanisms of D-amino acid oxidase inhibition by small molecules. J Med Chem. 2013 May 9;56(9):3710-24. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

