| Drug General Information |
| Drug ID |
D02JEZ
|
| Former ID |
DNC000103
|
| Drug Name |
7-hydroxycoumarin
|
| Drug Type |
Small molecular drug
|
| Structure |
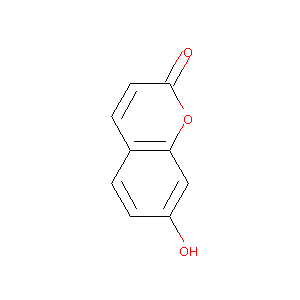
|
Download
2D MOL
3D MOL
|
| Formula |
C9H6O3
|
| Canonical SMILES |
C1=CC(=CC2=C1C=CC(=O)O2)O
|
| InChI |
1S/C9H6O3/c10-7-3-1-6-2-4-9(11)12-8(6)5-7/h1-5,10H
|
| InChIKey |
ORHBXUUXSCNDEV-UHFFFAOYSA-N
|
| PubChem Compound ID |
|
| PubChem Substance ID |
11506, 82592, 589071, 601312, 894672, 3133441, 6654657, 8138035, 8144499, 8616620, 10538273, 11371682, 11406039, 11485819, 11489778, 11490567, 11537968, 15321753, 24889947, 24895162, 26613209, 26679323, 26749818, 29204654, 35037624, 35037687, 36026715, 37041382, 39290237, 47365382, 47365383, 48334686, 49743161, 49835726, 49956270, 50123357, 50123358, 51074934, 53705440, 53789635, 57358161, 57391252, 57653583, 57968425, 81041515, 81092928, 85086760, 85789232, 85789383, 87550949
|
| Target and Pathway |
| References |