Drug Information
| Drug General Information | |||||
|---|---|---|---|---|---|
| Drug ID |
D06HRX
|
||||
| Former ID |
DNC003853
|
||||
| Drug Name |
AMENTOFLAVONE
|
||||
| Drug Type |
Small molecular drug
|
||||
| Indication | Discovery agent | Investigative | [526653] | ||
| Structure |
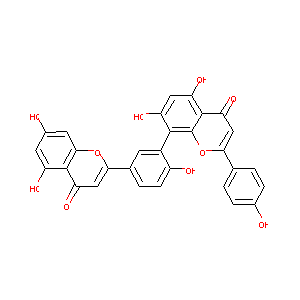
|
Download2D MOL |
|||
| Formula |
C30H18O10
|
||||
| Canonical SMILES |
C1=CC(=CC=C1C2=CC(=O)C3=C(O2)C(=C(C=C3O)O)C4=C(C=CC(=C4<br />)C5=CC(=O)C6=C(C=C(C=C6O5)O)O)O)O
|
||||
| InChI |
1S/C30H18O10/c31-15-4-1-13(2-5-15)24-12-23(38)29-21(36)10-20(35)27(30(29)40-24)17-7-14(3-6-18(17)33)25-11-22(37)28-19(34)8-16(32)9-26(28)39-25/h1-12,31-36H
|
||||
| InChIKey |
YUSWMAULDXZHPY-UHFFFAOYSA-N
|
||||
| PubChem Compound ID | |||||
| Target and Pathway | |||||
| Target(s) | Gamma-aminobutyric acid receptor subunit beta-2 | Target Info | Inhibitor | [526653] | |
| Induced myeloid leukemia cell differentiation protein Mcl-1 | Target Info | Inhibitor | [531262] | ||
| Gamma-aminobutyric-acid receptor alpha-5 subunit | Target Info | Inhibitor | [526653] | ||
| Gamma-aminobutyric acid receptor subunit alpha-1 | Target Info | Inhibitor | [526653] | ||
| Gamma-aminobutyric acid receptor subunit gamma-2 | Target Info | Inhibitor | [526653] | ||
| Gamma-aminobutyric-acid receptor alpha-2 subunit | Target Info | Inhibitor | [526653] | ||
| Gamma-aminobutyric-acid receptor alpha-3 subunit | Target Info | Inhibitor | [526653] | ||
| Xanthine dehydrogenase/oxidase | Target Info | Inhibitor | [551348] | ||
| Gamma-aminobutyric acid receptor | Target Info | Inhibitor | [526653] | ||
| KEGG Pathway | Neuroactive ligand-receptor interaction | ||||
| Retrograde endocannabinoid signaling | |||||
| Serotonergic synapse | |||||
| GABAergic synapse | |||||
| Morphine addiction | |||||
| Nicotine addictionhsa04151:PI3K-Akt signaling pathway | |||||
| MicroRNAs in cancerhsa04080:Neuroactive ligand-receptor interaction | |||||
| Nicotine addictionhsa04080:Neuroactive ligand-receptor interaction | |||||
| Nicotine addictionhsa00230:Purine metabolism | |||||
| Caffeine metabolism | |||||
| Drug metabolism - other enzymes | |||||
| Metabolic pathways | |||||
| Peroxisome | |||||
| NetPath Pathway | TCR Signaling Pathway | ||||
| Pathway Interaction Database | E2F transcription factor network | ||||
| Direct p53 effectors | |||||
| IL6-mediated signaling events | |||||
| HIF-1-alpha transcription factor network | |||||
| PathWhiz Pathway | Caffeine Metabolism | ||||
| Purine Metabolism | |||||
| WikiPathways | Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | ||||
| Iron uptake and transportWP254:Apoptosis | |||||
| miR-targeted genes in muscle cell - TarBase | |||||
| miR-targeted genes in lymphocytes - TarBase | |||||
| miR-targeted genes in leukocytes - TarBase | |||||
| Apoptosis Modulation and SignalingWP2754:Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | |||||
| Iron uptake and transportWP706:SIDS Susceptibility Pathways | |||||
| Iron uptake and transportWP2754:Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | |||||
| Iron uptake and transportWP408:Oxidative Stress | |||||
| Effects of Nitric Oxide | |||||
| Metabolism of nucleotides | |||||
| Selenium Micronutrient Network | |||||
| References | |||||
| Ref 526653 | Bioorg Med Chem Lett. 2003 Jul 21;13(14):2281-4.Semisynthetic preparation of amentoflavone: A negative modulator at GABA(A) receptors. | ||||
| Ref 531262 | Bioorg Med Chem Lett. 2010 Dec 15;20(24):7331-6. Epub 2010 Oct 21.In silico identification and biochemical evaluation of novel inhibitors of NRH:quinone oxidoreductase 2 (NQO2). | ||||
| Ref 551348 | Inhibition of cow's milk xanthine oxidase by flavonoids. J Nat Prod. 1988 Mar-Apr;51(2):345-8. | ||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

