Drug Information
| Drug General Information | |||||
|---|---|---|---|---|---|
| Drug ID |
D0SO9G
|
||||
| Former ID |
DNC011285
|
||||
| Drug Name |
5'-methylthio-immucillin-H
|
||||
| Indication | Discovery agent | Investigative | [531149] | ||
| Structure |
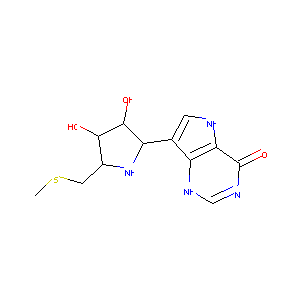
|
Download2D MOL |
|||
| Canonical SMILES |
CSCC1C(C(C(N1)C2=CNC3=C2NC=NC3=O)O)O
|
||||
| InChI |
1S/C12H16N4O3S/c1-20-3-6-10(17)11(18)8(16-6)5-2-13-9-7(5)14-4-15-12(9)19/h2,4,6,8,10-11,13,16-18H,3H2,1H3,(H,14,15,19)/t6-,8-,10-,11-/m0/s1
|
||||
| InChIKey |
CEGIKIXYDFDYDN-SEZOGAINSA-N
|
||||
| Target and Pathway | |||||
| Target(s) | Purine nucleoside phosphorylase | Target Info | Inhibitor | [531149] | |
| BioCyc Pathway | Arsenate detoxification I (glutaredoxin) | ||||
| Purine nucleotides degradation | |||||
| Urate biosynthesis/inosine 5'-phosphate degradation | |||||
| Purine deoxyribonucleosides degradation | |||||
| Purine ribonucleosides degradation to ribose-1-phosphate | |||||
| Guanosine nucleotides degradation | |||||
| Adenosine nucleotides degradation | |||||
| Superpathway of purine nucleotide salvage | |||||
| Adenine and adenosine salvage III | |||||
| Guanine and guanosine salvage | |||||
| NetPath Pathway | TCR Signaling Pathway | ||||
| EGFR1 Signaling Pathway | |||||
| PathWhiz Pathway | Purine Metabolism | ||||
| Nicotinate and Nicotinamide Metabolism | |||||
| Reactome | Purine salvage | ||||
| Purine catabolism | |||||
| References | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

