Drug Information
| Drug General Information | Top | |||
|---|---|---|---|---|
| Drug ID |
D0MM8N
|
|||
| Former ID |
DNAP001531
|
|||
| Drug Name |
Idebenone
|
|||
| Synonyms |
idebenone; 58186-27-9; Idebenona; Idebenonum; CV 2619; Idebenonum [Latin]; Idebenona [Spanish]; Idebenone [INN:JAN]; CV-2619; 2-(10-hydroxydecyl)-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione; UNII-HB6PN45W4J; BRN 2001459; 6-(10-Hydroxydecyl)-2,3-dimethoxy-5-methyl-1,4-benzoquinone; HB6PN45W4J; 2-(10-Hydroxydecyl)-5,6-dimethoxy-3-methyl-1,4-benzoquinone; CHEBI:31687; JGPMMRGNQUBGND-UHFFFAOYSA-N; 2,5-Cyclohexadiene-1,4-dione, 2-(10-hydroxydecyl)-5,6-dimethoxy-3-methyl-; NCGC00160514-01; AK161748
Click to Show/Hide
|
|||
| Drug Type |
Small molecular drug
|
|||
| Indication | Cognitive impairment [ICD-11: 6D71; ICD-10: F06.7] | Approved | [1] | |
| Friedreich's ataxia [ICD-11: 8A03.10] | Approved | [1] | ||
| Company |
Santhera Pharmaceuticals
|
|||
| Structure |
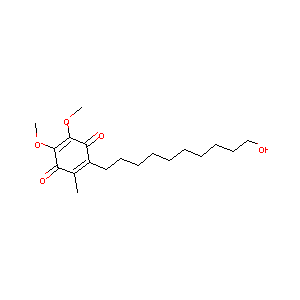 |
Download2D MOL |
||
| Formula |
C19H30O5
|
|||
| Canonical SMILES |
CC1=C(C(=O)C(=C(C1=O)OC)OC)CCCCCCCCCCO
|
|||
| InChI |
1S/C19H30O5/c1-14-15(12-10-8-6-4-5-7-9-11-13-20)17(22)19(24-3)18(23-2)16(14)21/h20H,4-13H2,1-3H3
|
|||
| InChIKey |
JGPMMRGNQUBGND-UHFFFAOYSA-N
|
|||
| CAS Number |
CAS 58186-27-9
|
|||
| PubChem Compound ID | ||||
| PubChem Substance ID |
1916788, 4725770, 7848813, 8152329, 12012810, 14802277, 26719872, 29216196, 29222809, 46386682, 49681642, 49957336, 56313750, 57321939, 78556875, 81041137, 81093345, 85279384, 88800993, 92308516, 92308744, 92710538, 103060386, 103556242, 104304229, 124658851, 125353623, 126631778, 126658121, 126665893, 128272043, 131319704, 131905204, 135003257, 135692548, 135698082, 136990616, 137027889, 142735616, 143492810, 144075938, 144205417, 152242914, 160871272, 162022270, 162038194, 162173185, 163079053, 164802349, 169618917
|
|||
| ChEBI ID |
CHEBI:31687
|
|||
| SuperDrug ATC ID |
N06BX13
|
|||
| Interaction between the Drug and Microbe | Top | |||
|---|---|---|---|---|
| The Metabolism of Drug Affected by Studied Microbe(s) | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Alistipes indistinctus DSM22520
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Alistipes indistinctus DSM22520 (log2FC = -0.622; p = 0.018). | |||
|
Studied Microbe: Bacteroides fragilis ATCC43859
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides fragilis ATCC43859 (log2FC = -0.799; p = 0.05). | |||
|
Studied Microbe: Bacteroides fragilis HMW 610
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides fragilis HMW 610 (log2FC = -0.988; p = 0.005). | |||
|
Studied Microbe: Bacteroides fragilis NCTC 9343
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides fragilis NCTC 9343 (log2FC = -0.876; p = 0.015). | |||
|
Studied Microbe: Bacteroides fragilis str. 3397 T10
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides fragilis str. 3397 T10 (log2FC = -0.636; p = 0.05). | |||
|
Studied Microbe: Bacteroides fragilis str. 3986 T(B)9
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides fragilis str. 3986 T(B)9 (log2FC = -0.765; p = 0.027). | |||
|
Studied Microbe: Bacteroides fragilis str. DS-208
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides fragilis str. DS-208 (log2FC = -0.706; p = 0.038). | |||
|
Studied Microbe: Bacteroides ovatus ATCC 8483
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides ovatus ATCC 8483 (log2FC = -0.357; p = 0.005). | |||
|
Studied Microbe: Bacteroides thetaiotaomicron 3731
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides thetaiotaomicron 3731 (log2FC = -0.464; p = 0.05). | |||
|
Studied Microbe: Bacteroides uniformis ATCC 8492
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides uniformis ATCC 8492 (log2FC = -0.839; p = 0.003). | |||
|
Studied Microbe: Bacteroides vulgatus ATCC 8482
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides vulgatus ATCC 8482 (log2FC = -0.708; p = 0.021). | |||
|
Studied Microbe: Bacteroides xylanisolvens DSM18836
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides xylanisolvens DSM18836 (log2FC = -0.782; p = 0.026). | |||
|
Studied Microbe: Odoribacter splanchnicus
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Odoribacter splanchnicus (log2FC = -0.772; p = 0.012). | |||
|
Studied Microbe: Parabacteroides distasonis ATCC 8503
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Parabacteroides distasonis ATCC 8503 (log2FC = -0.399; p = 0.022). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Eubacteriales | ||||
|
Studied Microbe: Bacteroides pectinophilus ATCC 43243
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Bacteroides pectinophilus ATCC 43243 (log2FC = -0.332; p = 0.026). | |||
|
Studied Microbe: Blautia luti DSM 14534
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Blautia luti DSM 14534 (log2FC = -5.089; p = 0.006). | |||
|
Studied Microbe: Clostridium sp.
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Clostridium sp. (log2FC = -0.563; p = 0.041). | |||
|
Studied Microbe: Enterocloster bolteae ATCC BAA-613
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Enterocloster bolteae ATCC BAA-613 (log2FC = -0.96; p = 0.015). | |||
|
Studied Microbe: Roseburia intestinalis L1-82
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Roseburia intestinalis L1-82 (log2FC = -0.364; p = 0.008). | |||
|
Studied Microbe: Ruminococcus lactaris ATCC 29176
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Ruminococcus lactaris ATCC 29176 (log2FC = -0.906; p = 0.044). | |||
|
Studied Microbe: Subdoligranulum variabile DSM 15176
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Idebenone can be metabolized by Subdoligranulum variabile DSM 15176 (log2FC = -0.7; p = 0.023). | |||
| The Abundace of Studied Microbe(s) Regulated by Drug | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Parabacteroides distasonis
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Parabacteroides distasonis was decreased by Idebenone (adjusted p-values: 3.82E-03). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Erysipelotrichales | ||||
|
Studied Microbe: Erysipelatoclostridium ramosum
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Erysipelatoclostridium ramosum was decreased by Idebenone (adjusted p-values: 6.61E-04). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Eubacteriales | ||||
|
Studied Microbe: Clostridium perfringens
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Clostridium perfringens was decreased by Idebenone (adjusted p-values: 4.61E-03). | |||
| References | Top | |||
|---|---|---|---|---|
| REF 1 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||
| REF 2 | Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019 Jun;570(7762):462-467. | |||
| REF 3 | Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018 Mar 29;555(7698):623-628. | |||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

