Drug Information
| Drug General Information | Top | |||
|---|---|---|---|---|
| Drug ID |
D08USJ
|
|||
| Former ID |
DAP000563
|
|||
| Drug Name |
Neostigmine
|
|||
| Synonyms |
Eustigmin; Eustigmine; Intrastigmina; Juvastigmin; Neostigmin; Neostigminum; Prostigmin; Prostigmine; Prozerin; Sulfonatooxymethane; Synstigmin; Syntostigmine; Vagostigmin; Vagostigmine; Neostigmine [BAN]; Neostigmine omega; M-Trimethylammoniumphenyldimethylcarbamate; Neostigmine (BAN); Prostigmin (TN); Vagostigmin (TN); [3-(dimethylcarbamoyloxy)phenyl]-trimethylazanium; [3-(dimethylcarbamoyloxy)phenyl]-trimethyl-azanium; [3-(dimethylcarbamoyloxy)phenyl]-trimethyl-azanium bromide; Ammonium, (m-hydroxyphenyl)trimethyl-, dimethylcarbamate (ester); Benzenaminium, 3-(((dimethylamino)carbonyl)oxy)-N,N,N-trimethyl-(9CI); (m-Hydroxyphenyl)trimethylammonium dimethylcarbamate; (m-Hydroxyphenyl)trimethylammonium dimethylcarbamate (ester); 3-Trimethylammoniumphenyl N,N-dimethylcarbamate; 3-[(dimethylcarbamoyl)oxy]-N,N,N-trimethylanilinium; 3-{[(dimethylamino)carbonyl]oxy}-N,N,N-trimethylanilinium
Click to Show/Hide
|
|||
| Drug Type |
Small molecular drug
|
|||
| Indication | Myasthenia gravis [ICD-11: 8C6Y; ICD-10: G70.0; ICD-9: 358] | Approved | [1] | |
| Therapeutic Class |
Parasympathomimetics
|
|||
| Structure |
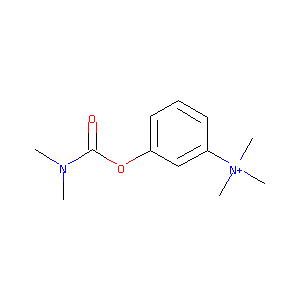 |
Download2D MOL |
||
| Formula |
C12H19N2O2+
|
|||
| Canonical SMILES |
CN(C)C(=O)OC1=CC=CC(=C1)[N+](C)(C)C
|
|||
| InChI |
1S/C12H19N2O2/c1-13(2)12(15)16-11-8-6-7-10(9-11)14(3,4)5/h6-9H,1-5H3/q+1
|
|||
| InChIKey |
ALWKGYPQUAPLQC-UHFFFAOYSA-N
|
|||
| CAS Number |
CAS 59-99-4
|
|||
| PubChem Compound ID | ||||
| PubChem Substance ID |
9467, 866380, 4398035, 4635954, 7980106, 8152746, 11111527, 11111528, 11335340, 11335373, 11360579, 11360612, 11363955, 11366517, 11369079, 11373762, 11377241, 11461551, 11461584, 11466496, 11467616, 11485012, 11486251, 11489094, 11491887, 11494875, 15121488, 26756597, 29223551, 46509161, 47216650, 47291007, 47365046, 47588860, 47736334, 47736335, 47810618, 48110324, 48416319, 49698982, 49868416, 50100289, 50100290, 50103950, 50765645, 53786926, 57322279, 81044623, 85787484, 87691541
|
|||
| ChEBI ID |
CHEBI:7514
|
|||
| ADReCS Drug ID | BADD_D01553 ; BADD_D01554 | |||
| SuperDrug ATC ID |
N07AA01; S01EB06
|
|||
| SuperDrug CAS ID |
cas=000059994
|
|||
| Interaction between the Drug and Microbe | Top | |||
|---|---|---|---|---|
| The Metabolism of Drug Affected by Studied Microbe(s) | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Bacteroides coprophilus DSM 18228
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides coprophilus DSM 18228 (log2FC = -0.533; p = 0.006). | |||
|
Studied Microbe: Bacteroides dorei DSM 17855
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides dorei DSM 17855 (log2FC = -0.528; p = 0.003). | |||
|
Studied Microbe: Bacteroides fragilis NCTC 9343
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides fragilis NCTC 9343 (log2FC = -0.611; p = 0.007). | |||
|
Studied Microbe: Bacteroides fragilis str. 3397 T10
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides fragilis str. 3397 T10 (log2FC = -0.693; p = 0.003). | |||
|
Studied Microbe: Bacteroides fragilis str. 3986 T(B)9
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides fragilis str. 3986 T(B)9 (log2FC = -0.535; p = 0.011). | |||
|
Studied Microbe: Bacteroides fragilis str. DS-208
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides fragilis str. DS-208 (log2FC = -0.584; p = 0.013). | |||
|
Studied Microbe: Bacteroides ovatus ATCC 8483
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides ovatus ATCC 8483 (log2FC = -0.62; p = 0.003). | |||
|
Studied Microbe: Bacteroides thetaiotaomicron 3731
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides thetaiotaomicron 3731 (log2FC = -0.352; p = 0.009). | |||
|
Studied Microbe: Bacteroides thetaiotaomicron 7330
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides thetaiotaomicron 7330 (log2FC = -0.598; p = 0.035). | |||
|
Studied Microbe: Bacteroides thetaiotaomicron VPI-5482
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides thetaiotaomicron VPI-5482 (log2FC = -0.515; p = 0.008). | |||
|
Studied Microbe: Bacteroides uniformis ATCC 8492
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Bacteroides uniformis ATCC 8492 (log2FC = -0.568; p = 0.001). | |||
|
Studied Microbe: Parabacteroides merdae ATCC 43184
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Neostigmine bromide can be metabolized by Parabacteroides merdae ATCC 43184 (log2FC = -0.342; p = 0.003). | |||
| Target and Pathway | Top | |||
|---|---|---|---|---|
| Target(s) | Acetylcholinesterase (AChE) | Target Info | Inhibitor | [3], [4] |
| KEGG Pathway | Glycerophospholipid metabolism | |||
| Cholinergic synapse | ||||
| Panther Pathway | Muscarinic acetylcholine receptor 1 and 3 signaling pathway | |||
| Muscarinic acetylcholine receptor 2 and 4 signaling pathway | ||||
| Nicotinic acetylcholine receptor signaling pathway | ||||
| Pathwhiz Pathway | Phospholipid Biosynthesis | |||
| Pathway Interaction Database | ATF-2 transcription factor network | |||
| WikiPathways | Monoamine Transport | |||
| Biogenic Amine Synthesis | ||||
| Acetylcholine Synthesis | ||||
| Integrated Pancreatic Cancer Pathway | ||||
| References | Top | |||
|---|---|---|---|---|
| REF 1 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||
| REF 2 | Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019 Jun;570(7762):462-467. | |||
| REF 3 | Screening of acetylcholinesterase inhibitors by CE after enzymatic reaction at capillary inlet. J Sep Sci. 2009 May;32(10):1748-56. | |||
| REF 4 | Neuromuscular blockade, reversal agent use, and operating room time: retrospective analysis of US inpatient surgeries. Curr Med Res Opin. 2009 Apr;25(4):943-50. | |||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

