Target expression details
| Target's General Information | |||||
|---|---|---|---|---|---|
| Target ID | T41560 | ||||
| Target Name | EGFR messenger RNA (EGFR mRNA) | ||||
| Synonyms | Receptor tyrosine-protein kinase erbB-1 (mRNA); Proto-oncogene c-ErbB-1 (mRNA); HER1 (mRNA); Epidermal growth factor receptor (mRNA); ERBB1 (mRNA); ERBB (mRNA); EGFR (mRNA) | ||||
| Target Type | Clinical trial | ||||
| Gene Name | EGFR | ||||
| Biochemical Class | mRNA target | ||||
| UniProt ID | EGFR_HUMAN | ||||
| Target's Expression Profile in Disease Related Tissue between Patients and Normal People | |||||
| Disease | Head and neck cancer | ||||
| Example drug | EGFR antisense DNA | Phase 1/2 | [1], [2], [3] | ||
| Tissue | Head and neck tissue | ||||
| Level of differential expression between the patients in the disease section of the tissue and the tissues of healthy individual |
Fold-change: 0.32 Z-score: 0.55 P-value: 5.84E-05 |
||||
|
Target gene expression profiles of the patients in the disease section of the tissue
Target gene expression profiles in the tissue of healthy individual
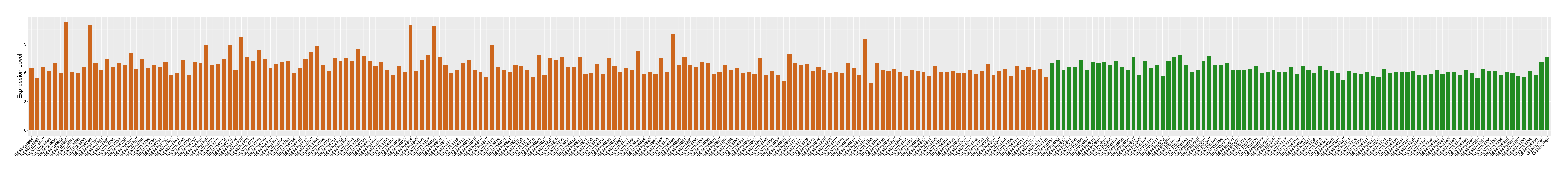
|
|||||
| Disease | Breast cancer | ||||
| Tissue | Breast tissue | ||||
| Level of differential expression between the patients in the disease section of the tissue and the tissues of healthy individual |
Fold-change: -0.89 Z-score: -1.53 P-value: 1.04E-65 |
||||
| Level of differential expression between the patients in the disease section of the tissue section of the tissue adjacent to the disease section |
Fold-change: -0.95 Z-score: -1.79 P-value: 2.77E-16 |
||||
|
Target gene expression profiles of the patients in the disease section of the tissue
Target gene expression profiles of the patients in the normal section of the tissue adjacent to the disease section
Target gene expression profiles in the tissue of healthy individual

|
|||||
| Disease | Esophagal cancer | ||||
| Tissue | Esophagus | ||||
| Level of differential expression between the patients in the disease section of the tissue section of the tissue adjacent to the disease section |
Fold-change: -0.93 Z-score: -1.08 P-value: 7.44E-01 |
||||
|
Target gene expression profiles of the patients in the disease section of the tissue
Target gene expression profiles of the patients in the normal section of the tissue adjacent to the disease section
Target gene expression profiles in the tissue of healthy individual
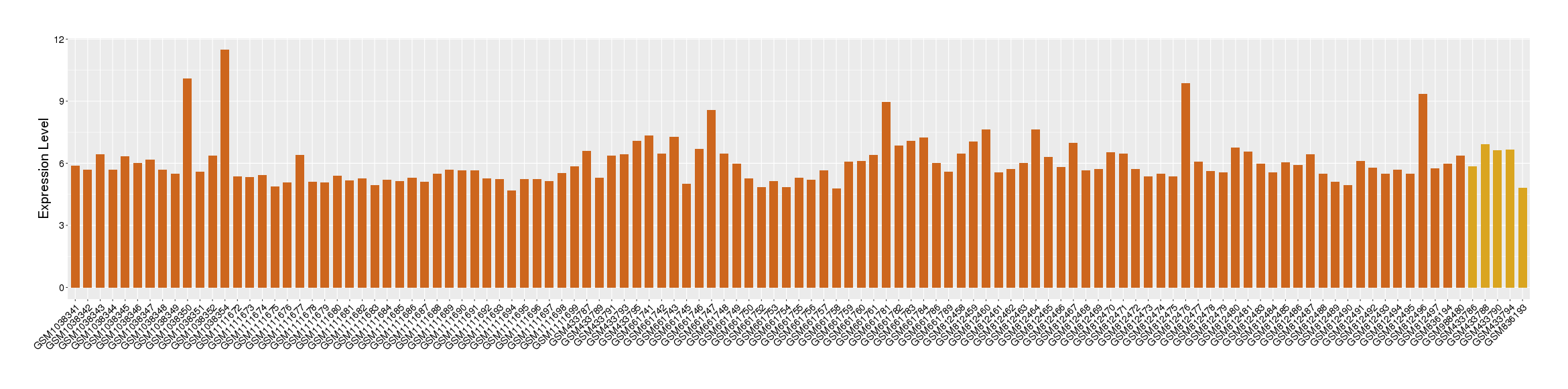
|
|||||
| Disease | Lung cancer | ||||
| Tissue | Lung tissue | ||||
| Level of differential expression between the patients in the disease section of the tissue and the tissues of healthy individual |
Fold-change: -0.04 Z-score: -0.09 P-value: 1.65E-01 |
||||
| Level of differential expression between the patients in the disease section of the tissue section of the tissue adjacent to the disease section |
Fold-change: -0.21 Z-score: -0.35 P-value: 1.58E-03 |
||||
|
Target gene expression profiles of the patients in the disease section of the tissue
Target gene expression profiles of the patients in the normal section of the tissue adjacent to the disease section
Target gene expression profiles in the tissue of healthy individual

|
|||||
| Disease | Psoriasis | ||||
| Tissue | Skin | ||||
| Level of differential expression between the patients in the disease section of the tissue and the tissues of healthy individual |
Fold-change: -0.27 Z-score: -0.42 P-value: 1.86E-07 |
||||
| Level of differential expression between the patients in the disease section of the tissue section of the tissue adjacent to the disease section |
Fold-change: 1.02 Z-score: 1.93 P-value: 1.31E-21 |
||||
|
Target gene expression profiles of the patients in the disease section of the tissue
Target gene expression profiles of the patients in the normal section of the tissue adjacent to the disease section
Target gene expression profiles in the tissue of healthy individual

|
|||||
| Target's Expression Profile across Various Tissues of Healthy Individual | |||||
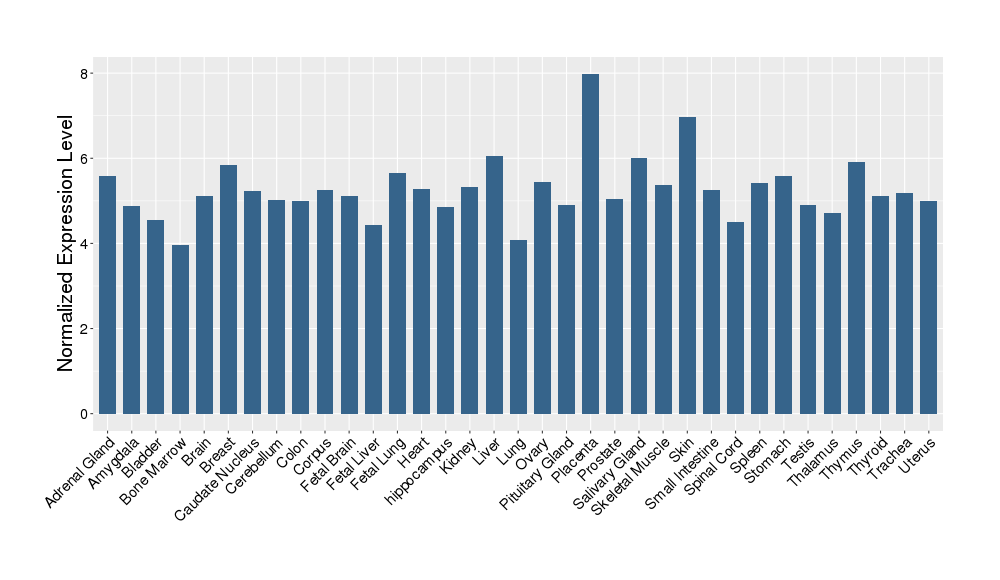
|
|||||
| References | |||||
| REF 1 | ClinicalTrials.gov (NCT00903461) Safety and Efficacy of Radiation/Cetuximab Plus EGFR Antisense DNA for Head and Neck Squamous Cell Carcinoma. U.S. National Institutes of Health. | ||||
| REF 2 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | ||||
| REF 3 | NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013 Jan;41(Database issue):D991-5. | ||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

