Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T99089
(Former ID: TTDS00346)
|
|||||
| Target Name |
Serine/threonine-protein kinase B-raf (BRAF)
|
|||||
| Synonyms |
V-Raf murine sarcoma viral oncogene homolog B1; RAFB1; Proto-oncogene B-Raf; P94; BRAF1; BRAF(V599E); BRAF serine/threonine kinase; B-raf protein; B-Raf
Click to Show/Hide
|
|||||
| Gene Name |
BRAF
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Melanoma [ICD-11: 2C30] | |||||
| Function |
May play a role in the postsynaptic responses of hippocampal neuron. Phosphorylates MAP2K1, and thereby contributes to the MAP kinase signal transduction pathway. Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MAALSGGGGGGAEPGQALFNGDMEPEAGAGAGAAASSAADPAIPEEVWNIKQMIKLTQEH
IEALLDKFGGEHNPPSIYLEAYEEYTSKLDALQQREQQLLESLGNGTDFSVSSSASMDTV TSSSSSSLSVLPSSLSVFQNPTDVARSNPKSPQKPIVRVFLPNKQRTVVPARCGVTVRDS LKKALMMRGLIPECCAVYRIQDGEKKPIGWDTDISWLTGEELHVEVLENVPLTTHNFVRK TFFTLAFCDFCRKLLFQGFRCQTCGYKFHQRCSTEVPLMCVNYDQLDLLFVSKFFEHHPI PQEEASLAETALTSGSSPSAPASDSIGPQILTSPSPSKSIPIPQPFRPADEDHRNQFGQR DRSSSAPNVHINTIEPVNIDDLIRDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSP GPQRERKSSSSSEDRNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDV AVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHH LHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATV KSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNIN NRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARS LPKIHRSASEPSLNRAGFQTEDFSLYACASPKTPIQAGGYGAFPVH Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T59C5R | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 3 Approved Drugs | + | ||||
| 1 | Dabrafenib | Drug Info | Approved | Melanoma | [2] | |
| 2 | LGX818 | Drug Info | Approved | Melanoma | [3] | |
| 3 | Vemurafenib | Drug Info | Approved | Melanoma | [4], [5], [6] | |
| Clinical Trial Drug(s) | [+] 10 Clinical Trial Drugs | + | ||||
| 1 | PLX8394 | Drug Info | Phase 2 | Solid tumour/cancer | [7] | |
| 2 | BGB-283 | Drug Info | Phase 1/2 | Solid tumour/cancer | [8] | |
| 3 | CEP-32496 | Drug Info | Phase 1/2 | Solid tumour/cancer | [9], [10] | |
| 4 | ABM-1310 | Drug Info | Phase 1 | Solid tumour/cancer | [11] | |
| 5 | ARQ 736 | Drug Info | Phase 1 | Solid tumour/cancer | [12] | |
| 6 | ASN003 | Drug Info | Phase 1 | Solid tumour/cancer | [7] | |
| 7 | BMS-908662 | Drug Info | Phase 1 | Solid tumour/cancer | [13], [14] | |
| 8 | PF-07284890 | Drug Info | Phase 1 | Melanoma | [15] | |
| 9 | PF-07799933 | Drug Info | Phase 1 | Aggressive cancer | [16] | |
| 10 | RO-5212054 | Drug Info | Phase 1 | Solid tumour/cancer | [17] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | RG-7256 | Drug Info | Discontinued in Phase 1 | Melanoma | [18] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 13 Inhibitor drugs | + | ||||
| 1 | Dabrafenib | Drug Info | [1], [19] | |||
| 2 | PLX8394 | Drug Info | [21] | |||
| 3 | BGB-283 | Drug Info | [22] | |||
| 4 | CEP-32496 | Drug Info | [23] | |||
| 5 | ABM-1310 | Drug Info | [24] | |||
| 6 | ASN003 | Drug Info | [7] | |||
| 7 | BMS-908662 | Drug Info | [14] | |||
| 8 | PF-07284890 | Drug Info | [26] | |||
| 9 | PF-07799933 | Drug Info | [16] | |||
| 10 | RO-5212054 | Drug Info | [27] | |||
| 11 | RG-7256 | Drug Info | [28] | |||
| 12 | AR-00341677 | Drug Info | [29] | |||
| 13 | AZ-628 | Drug Info | [30] | |||
| Modulator | [+] 3 Modulator drugs | + | ||||
| 1 | LGX818 | Drug Info | [20] | |||
| 2 | Vemurafenib | Drug Info | [5] | |||
| 3 | ARQ 736 | Drug Info | [25] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Ponatinib | Ligand Info | |||||
| Structure Description | The co-crystal structure of BRAF(V600E) with ponatinib | PDB:6P3D | ||||
| Method | X-ray diffraction | Resolution | 2.11 Å | Mutation | No | [31] |
| PDB Sequence |
KMDDWEIPDG
455 QITVGQRIGS465 GSFGTVYKGK475 WHGDVAVKML485 NVTAPTPQQL495 QAFKNEVGVL 505 RKTRHVNILL515 FMGYSTKPQL525 AIVTQWCEGS535 SLYHHLHASE545 TKFEMKKLID 555 IARQTARGMD565 YLHAKSIIHR575 DLKSNNIFLH585 EDNTVKIGDF595 GLATESGSIL 618 WMAPEVIRMQ628 DSNPYSFQSD638 VYAFGIVLYE648 LMTGQLPYSN658 INNRDQIIEM 668 VGRGSLSPDL678 SKVRSNCPKR688 MKRLMAECLK698 KKRDERPSFP708 RILAEIEELA 718 RELS
|
|||||
|
|
ILE463
3.957
VAL471
3.653
ALA481
3.280
VAL482
3.769
LYS483
3.415
GLU501
2.724
VAL504
4.004
LEU505
3.612
THR508
4.350
ILE513
3.549
LEU514
3.237
ILE527
3.616
VAL528
4.920
THR529
3.392
GLN530
3.300
|
|||||
| Ligand Name: Dabrafenib | Ligand Info | |||||
| Structure Description | B-Raf Kinase V600E oncogenic mutant in complex with Dabrafenib | PDB:4XV2 | ||||
| Method | X-ray diffraction | Resolution | 2.50 Å | Mutation | Yes | [32] |
| PDB Sequence |
DWEIPDGQIT
458 VGQRIGSGSF468 GTVYKGKWHG478 DVAVKMLNVP490 TPQQLQAFKN500 EVGVLRKTRH 510 VNILLFMGYS520 TKPQLAIVTQ530 WCEGSSLYHH540 LHASETKFEM550 KKLIDIARQT 560 ARGMDYLHAK570 SIIHRDLKSN580 NIFLHEDNTV590 KIGDFGGSIL618 WMAPEVIRPY 633 SFQSDVYAFG643 IVLYELMTGQ653 LPYSNINNRD663 QIIEMVGRGS673 LSPDLSKVRS 683 NCPKRMKRLM693 AECLKKKRDE703 RPSFPRILAE713 IEELARE
|
|||||
|
|
ILE463
4.245
GLY464
3.646
SER465
3.673
GLY466
3.826
PHE468
3.667
VAL471
3.304
ALA481
3.614
LYS483
2.878
LEU505
3.291
THR508
4.773
ILE513
4.611
LEU514
3.116
LEU515
4.412
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| ErbB signaling pathway | hsa04012 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Chemokine signaling pathway | hsa04062 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| mTOR signaling pathway | hsa04150 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |

|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Natural killer cell mediated cytotoxicity | hsa04650 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Long-term potentiation | hsa04720 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Serotonergic synapse | hsa04726 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Long-term depression | hsa04730 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
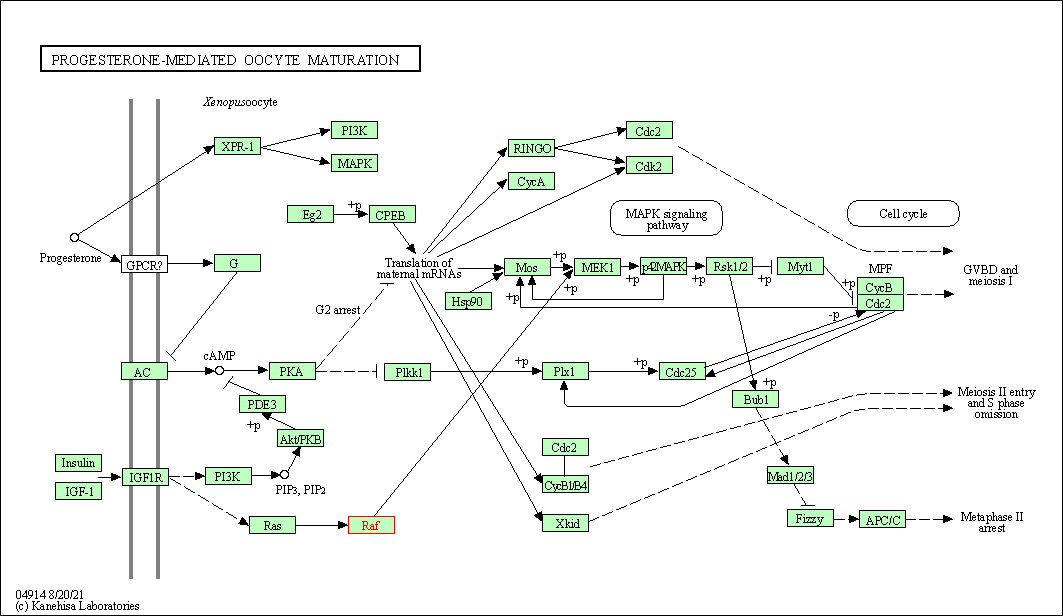
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
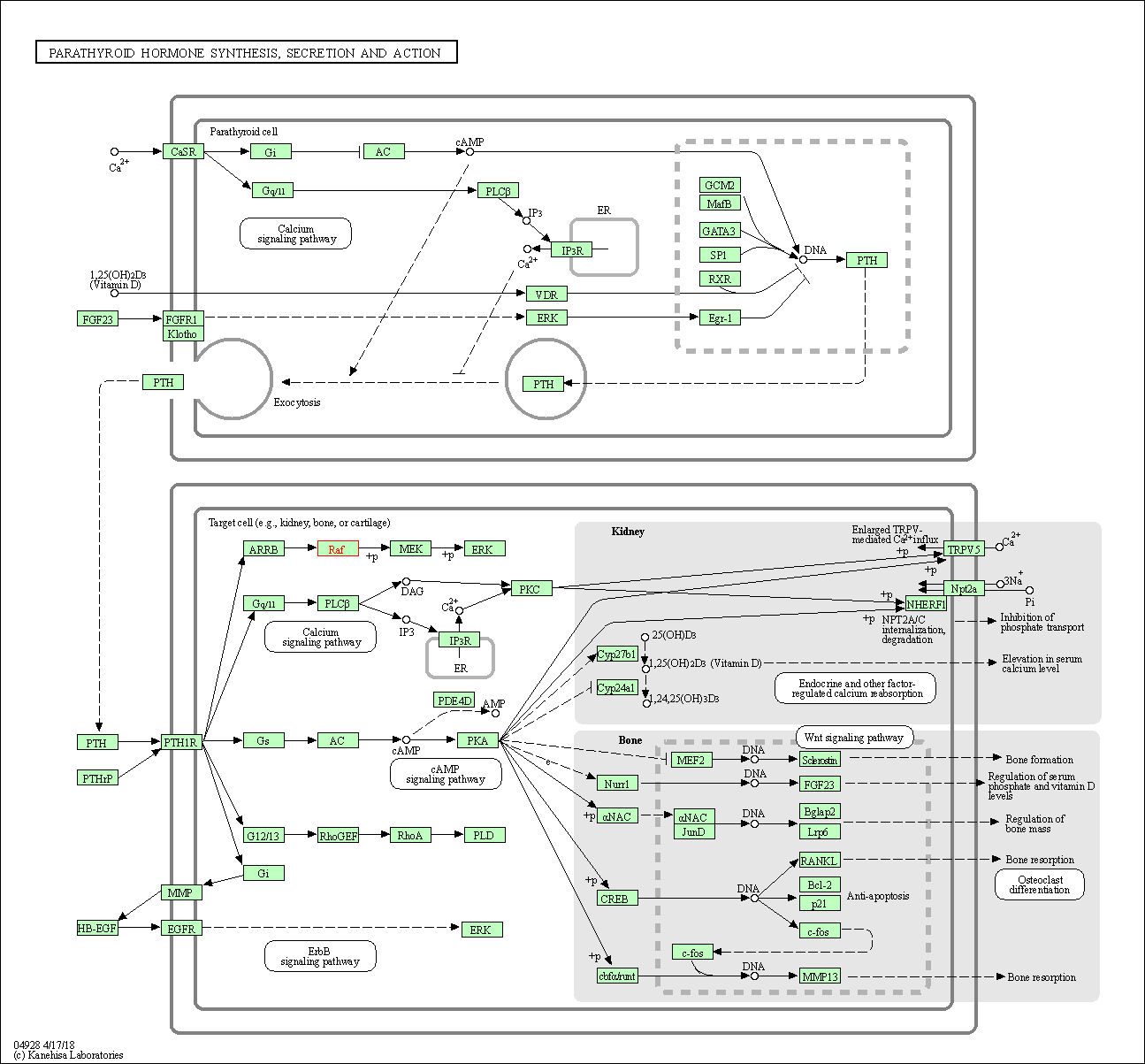
| Parathyroid hormone synthesis, secretion and action | hsa04928 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 21 | Degree centrality | 2.26E-03 | Betweenness centrality | 1.48E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.33E-01 | Radiality | 1.41E+01 | Clustering coefficient | 3.71E-01 |
| Neighborhood connectivity | 3.55E+01 | Topological coefficient | 1.16E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Drug Resistance Mutation (DRM) |
||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of GlaxoSmithKline (2011). | |||||
| REF 2 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. | |||||
| REF 3 | 2018 FDA drug approvals.Nat Rev Drug Discov. 2019 Feb;18(2):85-89. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5893). | |||||
| REF 5 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 6 | ClinicalTrials.gov (NCT01898585) An Open-Label Study of Zelboraf (Vemurafenib) in Patients With Braf V600 Mutation Positive Metastatic Melanoma. U.S. National Institutes of Health. | |||||
| REF 7 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 8 | ClinicalTrials.gov (NCT03905148) Study of the Safety and Pharmacokinetics of BGB-283 (Lifirfenib) and PD-0325901 (Mirdametinib) in Participants With Advanced or Refractory Solid Tumors. U.S. National Institutes of Health. | |||||
| REF 9 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7880). | |||||
| REF 10 | ClinicalTrials.gov (NCT01877811) CEP-32496 in Patients With Advanced Solid Tumors in Phase 1 and Advanced Melanoma and Metastatic Colorectal Cancer in Phase 2. U.S. National Institutes of Health. | |||||
| REF 11 | ClinicalTrials.gov (NCT04190628) Safety of ABM-1310 Monotherapy in Patients With Advanced Solid Tumors. U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT01225536) Dose Escalation Study of ARQ 736 in Adult Subjects With Advanced Solid Tumors Harboring BRAF and/or NRAS Mutations. U.S. National Institutes of Health. | |||||
| REF 13 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7968). | |||||
| REF 14 | Clinical pipeline report, company report or official report of Exelixis (2011). | |||||
| REF 15 | ClinicalTrials.gov (NCT04543188) A TWO-PART, PHASE 1A/B, OPEN-LABEL, MULTICENTER TRIAL EVALUATING PHARMACOKINETICS, SAFETY AND EFFICACY OF PF-07284890 (ARRY 461) IN PARTICIPANTS WITH BRAF V600 MUTANT SOLID TUMORS WITH AND WITHOUT BRAIN INVOLVEMENT. U.S.National Institutes of Health. | |||||
| REF 16 | ClinicalTrials.gov (NCT05355701) A PHASE 1, OPEN-LABEL, DOSE ESCALATION AND DOSE EXPANSION STUDY TO EVALUATE THE SAFETY, TOLERABILITY, PHARMACOKINETICS, AND ANTI TUMOR ACTIVITY OF PF-07799933 (ARRY-440) AS A SINGLE AGENT AND IN COMBINATION THERAPY IN PARTICIPANTS 16 YEARS AND OLDER WITH ADVANCED SOLID TUMORS WITH BRAF ALTERATIONS. U.S.National Institutes of Health. | |||||
| REF 17 | ClinicalTrials.gov (NCT01143753) A Study of RO5212054 (PLX3603) in Patients With BRAF V600-mutated Advanced Solid Tumours. U.S. National Institutes of Health. | |||||
| REF 18 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800022593) | |||||
| REF 19 | Caspase inhibitors: a review of recently patented compounds (2013-2015).Expert Opin Ther Pat. 2018 Jan;28(1):47-59. | |||||
| REF 20 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 21 | Identification of PLX4032-resistance mechanisms and implications for novel RAF inhibitors. Pigment Cell Melanoma Res. 2014 Mar;27(2):253-62. | |||||
| REF 22 | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers. Mol Cancer Ther. 2015 Oct;14(10):2187-97. | |||||
| REF 23 | Clinical pipeline report, company report or official report of Ambit Biosciences. | |||||
| REF 24 | Clinical pipeline report, company report or official report of ABM Therapeutics. | |||||
| REF 25 | National Cancer Institute Drug Dictionary (drug id 688029). | |||||
| REF 26 | Clinical pipeline report, company report or official report of Pfzer | |||||
| REF 27 | National Cancer Institute Drug Dictionary (drug id 680347). | |||||
| REF 28 | First-in-Man Dose-Escalation Study of the Selective BRAF Inhibitor RG7256 in Patients with BRAF V600-Mutated Advanced Solid Tumors. Target Oncol. 2016 Apr;11(2):149-56. | |||||
| REF 29 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5681). | |||||
| REF 30 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 8475). | |||||
| REF 31 | Inhibitors of BRAF dimers using an allosteric site. Nat Commun. 2020 Sep 1;11(1):4370. | |||||
| REF 32 | RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015 Oct 22;526(7574):583-6. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

