Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T04894
(Former ID: TTDI00043)
|
|||||
| Target Name |
Liver and activation-regulated chemokine (CCL20)
|
|||||
| Synonyms |
Smallinducible cytokine A20; Small-inducible cytokine A20; SCYA20; Macrophage inflammatory protein 3 alpha; MIP3alpha; MIP3A; MIP-3-alpha; Liver and activationregulated chemokine; LARC; CCL20(270); CC motif chemokine 20; CC chemokine LARC; C-C motif chemokine 20; Betachemokine exodus1; Beta-chemokine exodus-1
Click to Show/Hide
|
|||||
| Gene Name |
CCL20
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Psoriatic arthritis [ICD-11: FA21] | |||||
| Function |
Signals through binding and activation of CCR6 and induces a strong chemotactic response and mobilization of intracellular calcium ions. The ligand-receptor pair CCL20-CCR6 is responsible for the chemotaxis of dendritic cells (DC), effector/memory T-cells and B-cells and plays an important role at skin and mucosal surfaces under homeostatic and inflammatory conditions, as well as in pathology, including cancer and various autoimmune diseases. CCL20 acts as a chemotactic factor that attracts lymphocytes and, slightly, neutrophils, but not monocytes. Involved in the recruitment of both the proinflammatory IL17 producing helper T-cells (Th17) and the regulatory T-cells (Treg) to sites of inflammation. Required for optimal migration of thymic natural regulatory T cells (nTregs) and DN1 early thymocyte progenitor cells. C-terminal processed forms have been shown to be equally chemotactically active for leukocytes. Positively regulates sperm motility and chemotaxis via its binding to CCR6 which triggers Ca2+ mobilization in the sperm which is important for its motility. Inhibits proliferation of myeloid progenitors in colony formation assays. May be involved in formation and function of the mucosal lymphoid tissues by attracting lymphocytes and dendritic cells towards epithelial cells. Possesses antibacterial activity towards E. coli ATCC 25922 and S. aureus ATCC 29213. Acts as a ligand for C-C chemokine receptor CCR6.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cytokine: CC chemokine
|
|||||
| UniProt ID | ||||||
| Sequence |
MCCTKSLLLAALMSVLLLHLCGESEAASNFDCCLGYTDRILHPKFIVGFTRQLANEGCDI
NAIIFHTKKKLSVCANPKQTWVKYIVRLLSKKVKNM Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T49NJW | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | GSK3050002 | Drug Info | Phase 1 | Psoriatic arthritis | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | GSK3050002 | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
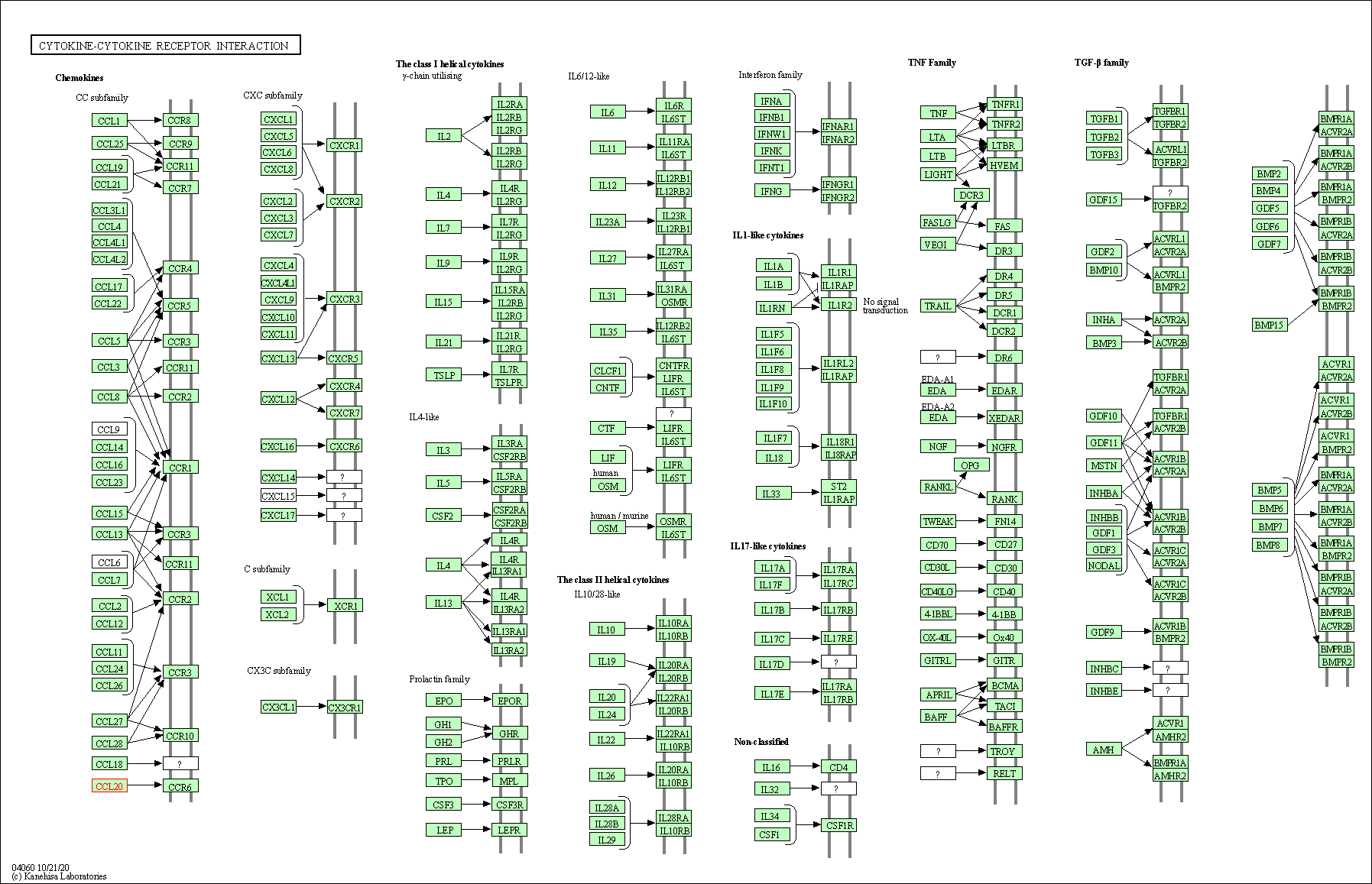
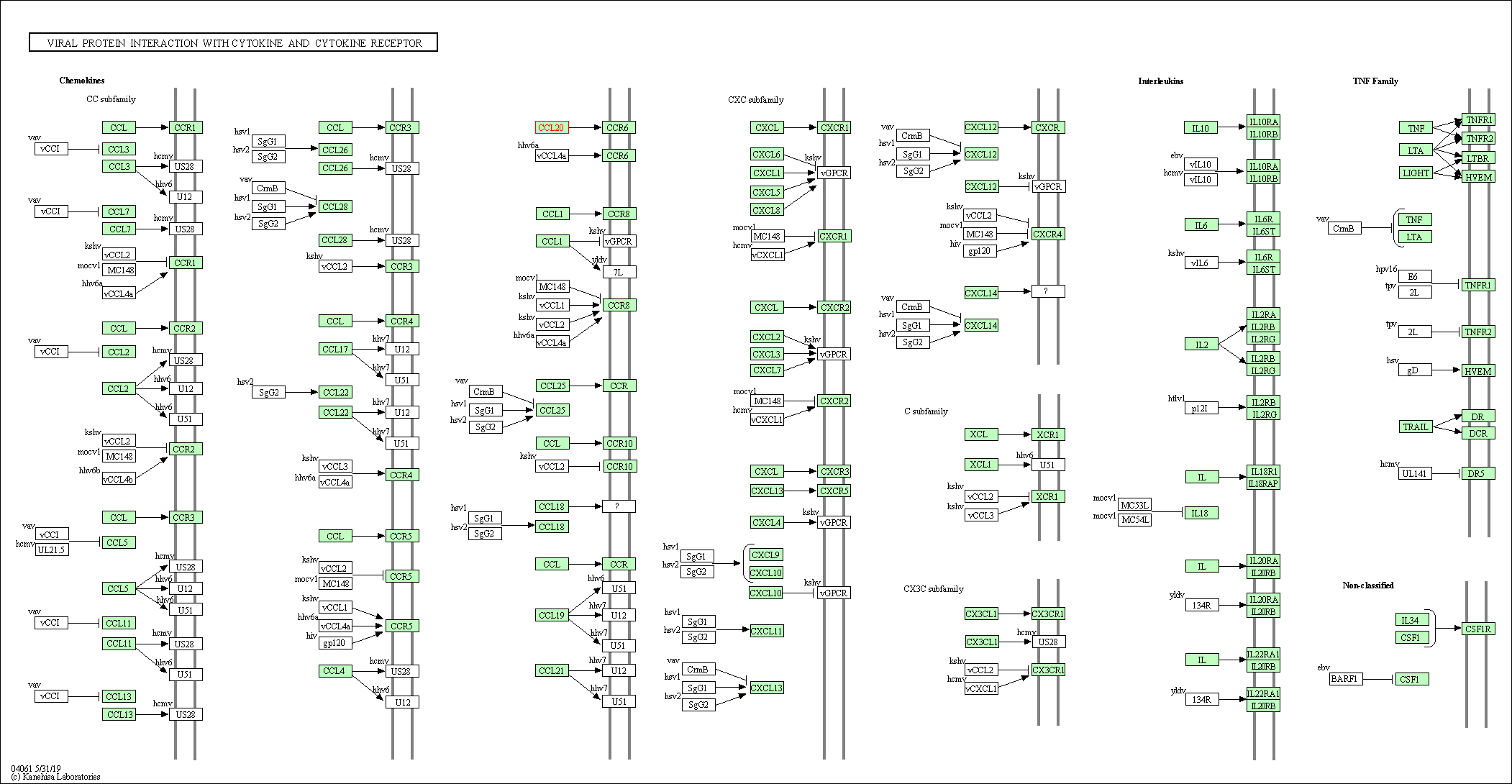
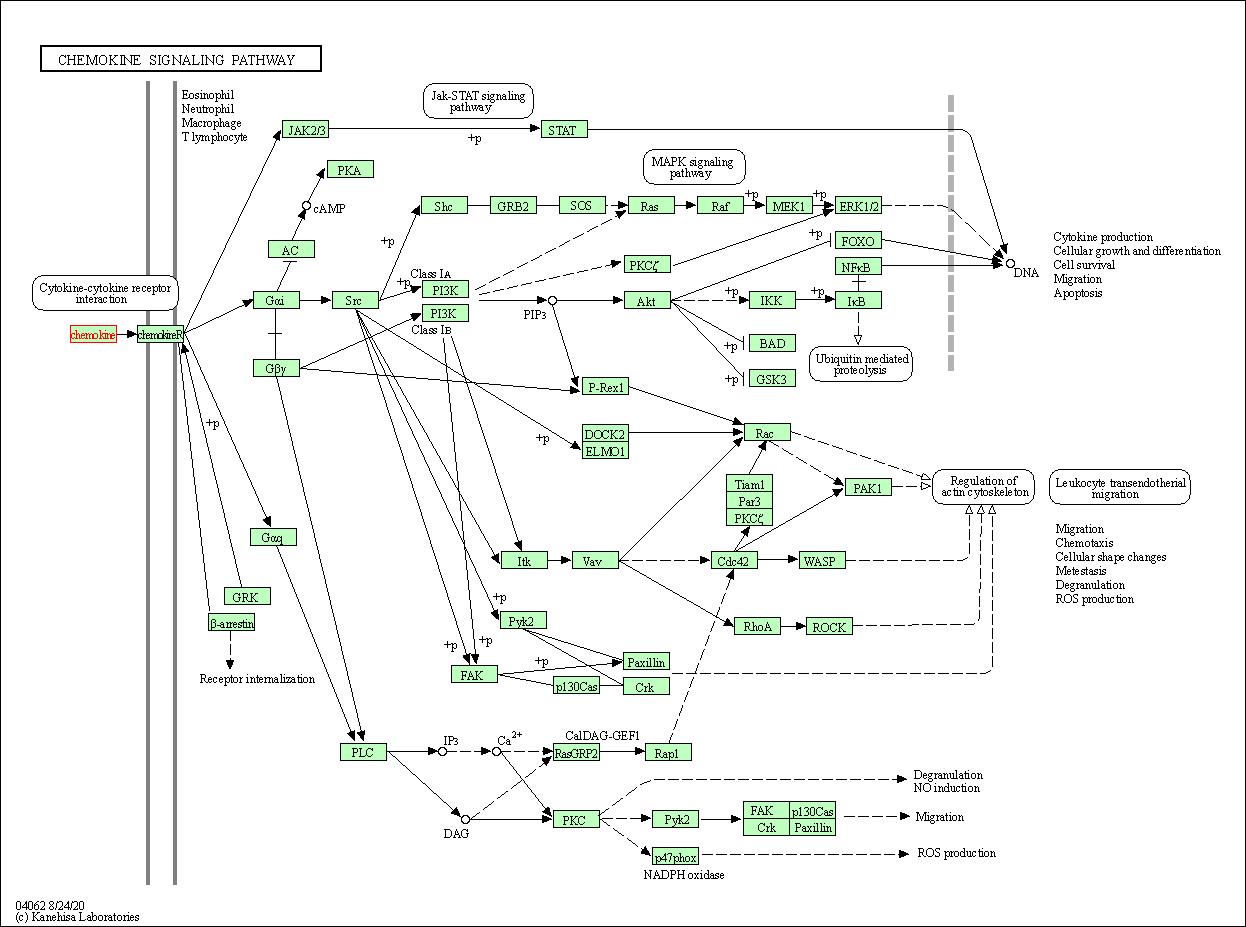
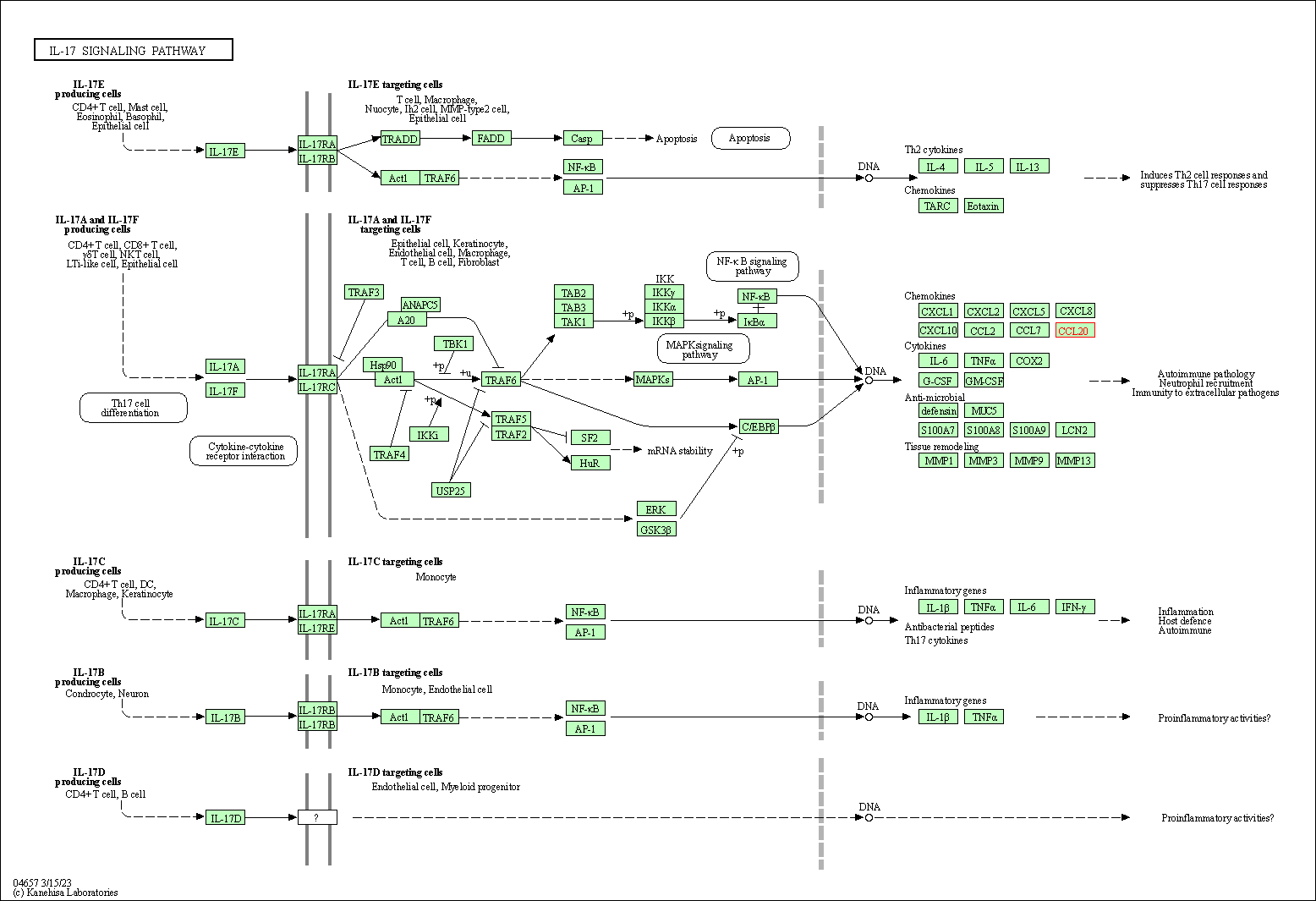
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | hsa04060 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Viral protein interaction with cytokine and cytokine receptor | hsa04061 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Chemokine signaling pathway | hsa04062 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
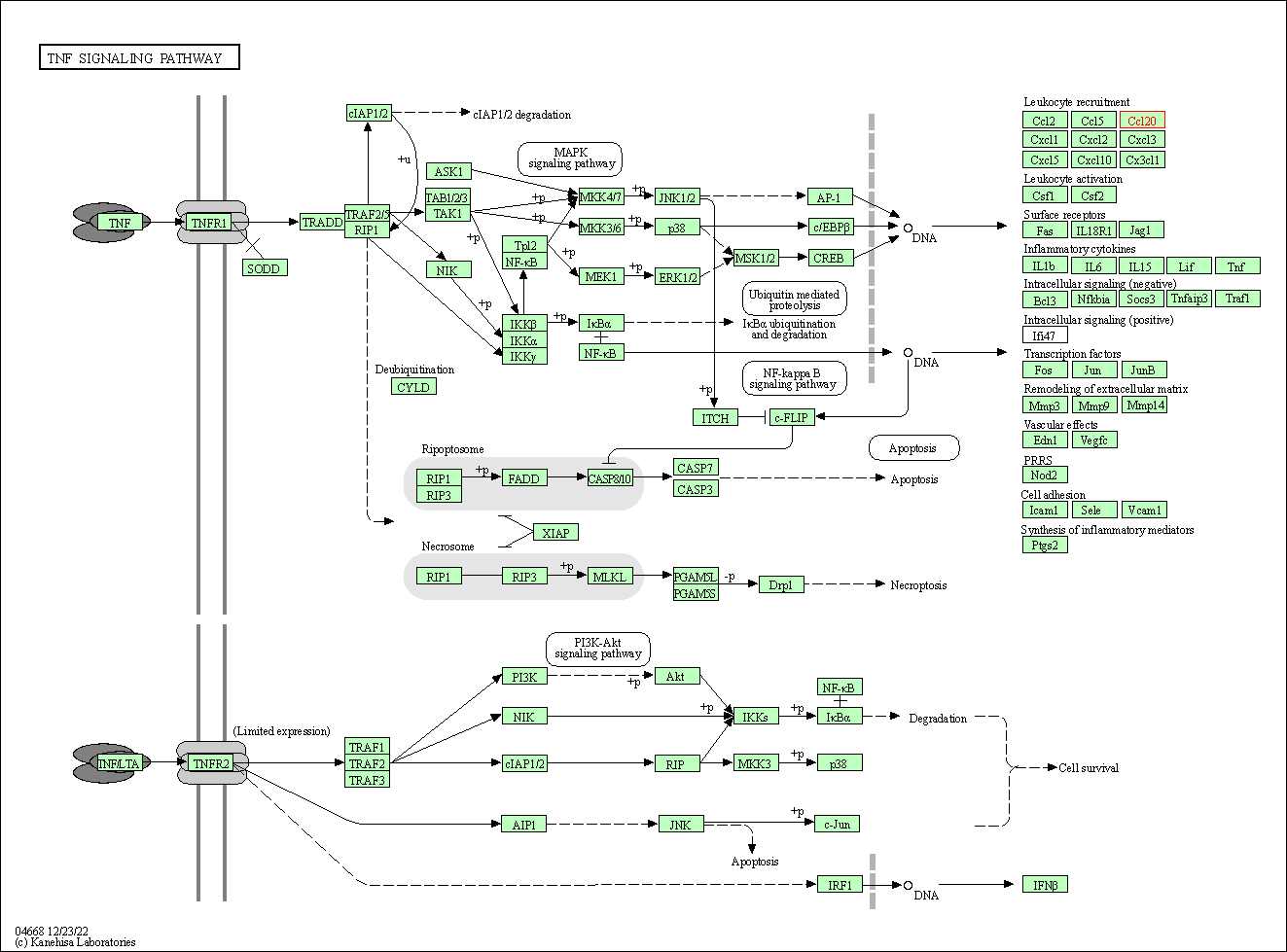
| TNF signaling pathway | hsa04668 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Degree | 14 | Degree centrality | 1.50E-03 | Betweenness centrality | 2.48E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.14E-01 | Radiality | 1.37E+01 | Clustering coefficient | 8.13E-01 |
| Neighborhood connectivity | 3.31E+01 | Topological coefficient | 1.97E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

