Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T04902
(Former ID: TTDR00505)
|
|||||
| Target Name |
Calpain-2 (CAPN2)
|
|||||
| Synonyms |
Millimolar-calpain; M-calpain; Calpain-2 large subunit; Calpain-2 catalytic subunit; Calpain large polypeptide L2; Calpain M-type; Calcium-activated neutral proteinase 2; Calcium-activated neutral proteinase; CANPL2; CANP 2; CANP
Click to Show/Hide
|
|||||
| Gene Name |
CAPN2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Alzheimer disease [ICD-11: 8A20] | |||||
| Function |
Proteolytically cleaves MYOC at 'Arg-226'. Proteolytically cleaves CPEB3 following neuronal stimulation which abolishes CPEB3 translational repressor activity, leading to translation of CPEB3 target mRNAs. Calcium-regulated non-lysosomal thiol-protease which catalyzes limited proteolysis of substrates involved in cytoskeletal remodeling and signal transduction.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.22.53
|
|||||
| Sequence |
MAGIAAKLAKDREAAEGLGSHDRAIKYLNQDYEALRNECLEAGTLFQDPSFPAIPSALGF
KELGPYSSKTRGIEWKRPTEICADPQFIIGGATRTDICQGALGDCWLLAAIASLTLNEEI LARVVPLNQSFQENYAGIFHFQFWQYGEWVEVVVDDRLPTKDGELLFVHSAEGSEFWSAL LEKAYAKINGCYEALSGGATTEGFEDFTGGIAEWYELKKPPPNLFKIIQKALQKGSLLGC SIDITSAADSEAITFQKLVKGHAYSVTGAEEVESNGSLQKLIRIRNPWGEVEWTGRWNDN CPSWNTIDPEERERLTRRHEDGEFWMSFSDFLRHYSRLEICNLTPDTLTSDTYKKWKLTK MDGNWRRGSTAGGCRNYPNTFWMNPQYLIKLEEEDEDEEDGESGCTFLVGLIQKHRRRQR KMGEDMHTIGFGIYEVPEELSGQTNIHLSKNFFLTNRARERSDTFINLREVLNRFKLPPG EYILVPSTFEPNKDGDFCIRVFSEKKADYQAVDDEIEANLEEFDISEDDIDDGFRRLFAQ LAGEDAEISAFELQTILRRVLAKRQDIKSDGFSIETCKIMVDMLDSDGSGKLGLKEFYIL WTKIQKYQKIYREIDVDRSGTMNSYEMRKALEEAGFKMPCQLHQVIVARFADDQLIIDFD NFVRCLVRLETLFKIFKQLDPENTGTIELDLISWLCFSVL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | ABT-957 | Drug Info | Phase 1 | Alzheimer disease | [2] | |
| Discontinued Drug(s) | [+] 4 Discontinued Drugs | + | ||||
| 1 | BITHIONOL | Drug Info | Withdrawn from market | Trematode infection | [3], [4] | |
| 2 | Aloxistatin | Drug Info | Discontinued in Phase 3 | Neurological disorder | [5] | |
| 3 | AK-295 | Drug Info | Terminated | Cerebral infarction | [6] | |
| 4 | MDL-28170 | Drug Info | Terminated | Alzheimer disease | [7] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | ABT-957 | Drug Info | [1] | |||
| Inhibitor | [+] 36 Inhibitor drugs | + | ||||
| 1 | 5-azolone derivative 1 | Drug Info | [8] | |||
| 2 | Carboxamide derivative 10 | Drug Info | [8] | |||
| 3 | Carboxamide derivative 11 | Drug Info | [8] | |||
| 4 | Carboxamide derivative 5 | Drug Info | [8] | |||
| 5 | Carboxamide derivative 6 | Drug Info | [8] | |||
| 6 | Carboxamide derivative 7 | Drug Info | [8] | |||
| 7 | Carboxamide derivative 8 | Drug Info | [8] | |||
| 8 | Carboxamide derivative 9 | Drug Info | [8] | |||
| 9 | Epoxysuccinate derivative 1 | Drug Info | [8] | |||
| 10 | Epoxysuccinate derivative 10 | Drug Info | [8] | |||
| 11 | Epoxysuccinate derivative 2 | Drug Info | [8] | |||
| 12 | Epoxysuccinate derivative 3 | Drug Info | [8] | |||
| 13 | Epoxysuccinate derivative 4 | Drug Info | [8] | |||
| 14 | Epoxysuccinate derivative 5 | Drug Info | [8] | |||
| 15 | Epoxysuccinate derivative 6 | Drug Info | [8] | |||
| 16 | Epoxysuccinate derivative 7 | Drug Info | [8] | |||
| 17 | Epoxysuccinate derivative 8 | Drug Info | [8] | |||
| 18 | Epoxysuccinate derivative 9 | Drug Info | [8] | |||
| 19 | PMID25399719-Compound-17 | Drug Info | [8] | |||
| 20 | BITHIONOL | Drug Info | [9] | |||
| 21 | Aloxistatin | Drug Info | [9] | |||
| 22 | AK-295 | Drug Info | [10] | |||
| 23 | MDL-28170 | Drug Info | [11] | |||
| 24 | SJA-6017 | Drug Info | [12] | |||
| 25 | (1-Benzyl-2-oxo-ethyl)-carbamic acid benzyl ester | Drug Info | [13] | |||
| 26 | 3-(1H-Pyrrol-3-yl)-propionamide | Drug Info | [14] | |||
| 27 | A-558693 | Drug Info | [10] | |||
| 28 | AW-00430 | Drug Info | [9] | |||
| 29 | Calpastatin | Drug Info | [15], [16] | |||
| 30 | DIHYDROXANTHOHUMOL | Drug Info | [9] | |||
| 31 | GNF-PF-4453 | Drug Info | [9] | |||
| 32 | mercaptoacrylate inhibitor of calpain 1 | Drug Info | [10] | |||
| 33 | N-(1-Benzyl-2-oxo-ethyl)-benzamide | Drug Info | [13] | |||
| 34 | PMID20690647C4b | Drug Info | [17] | |||
| 35 | PMID20690647C5a | Drug Info | [17] | |||
| 36 | PMID8831774C19 | Drug Info | [18] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
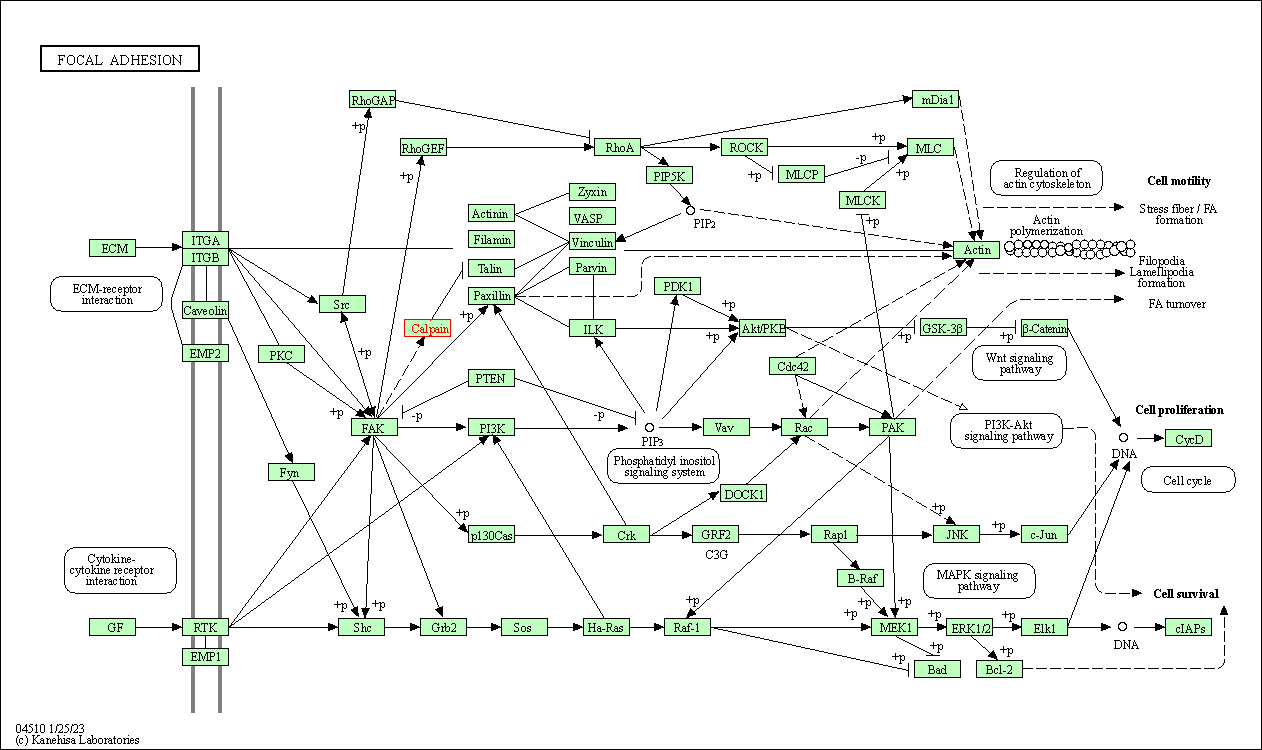
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
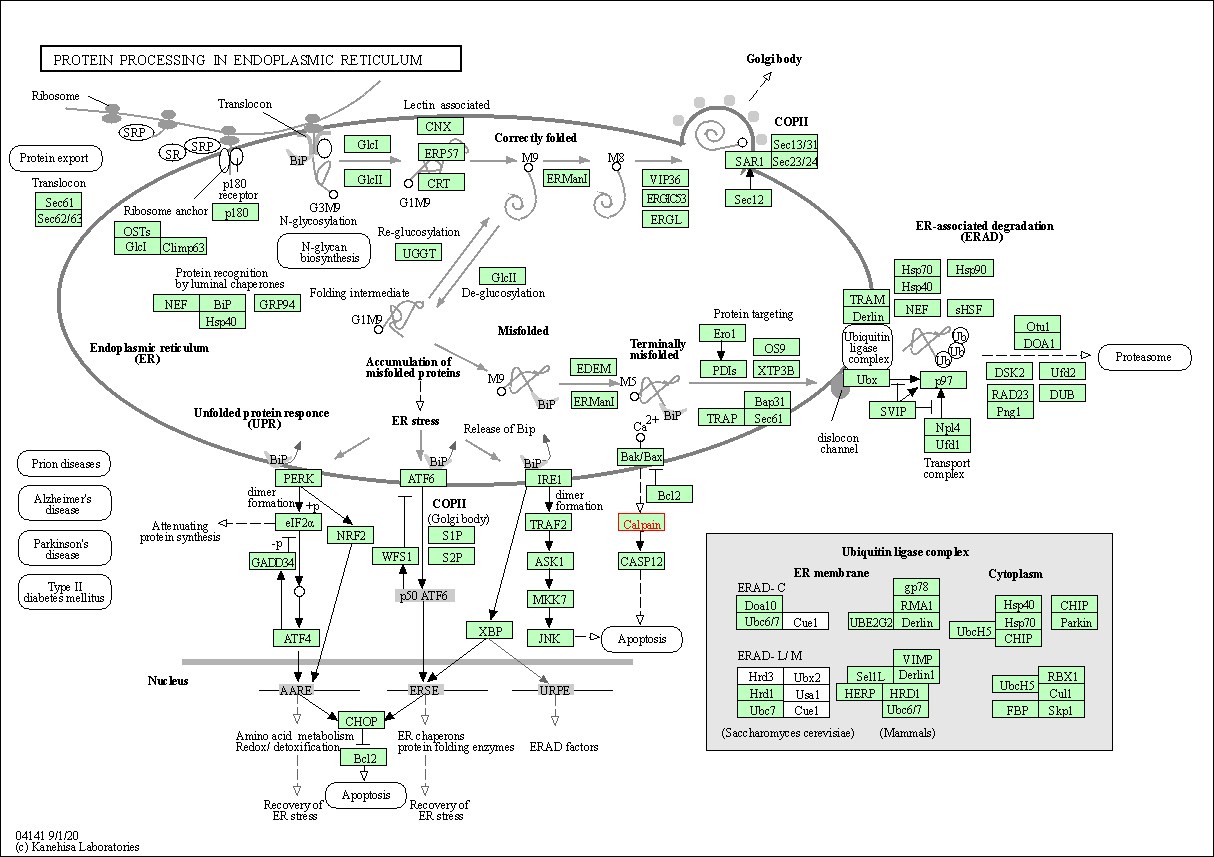
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |

|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
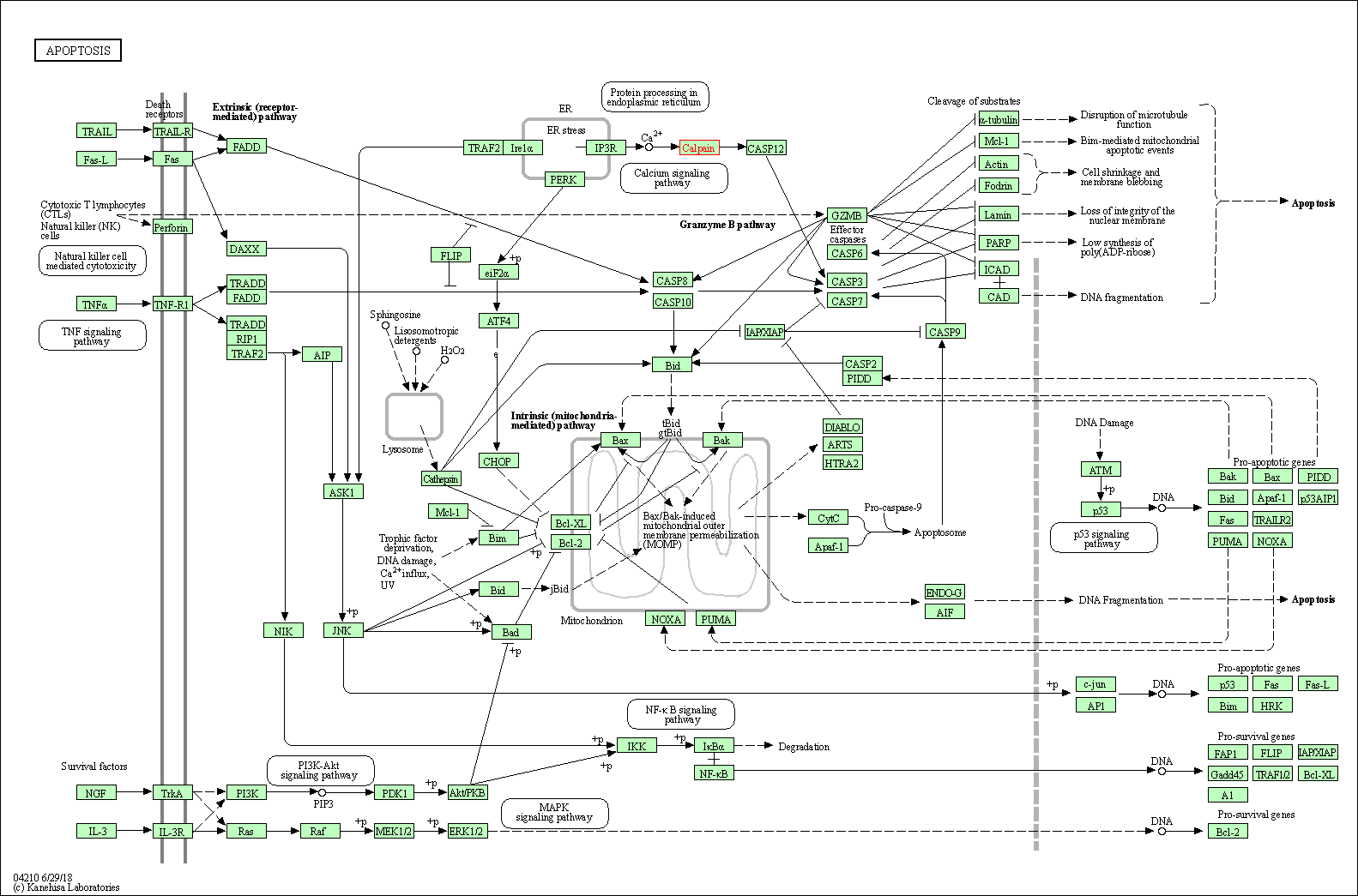
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
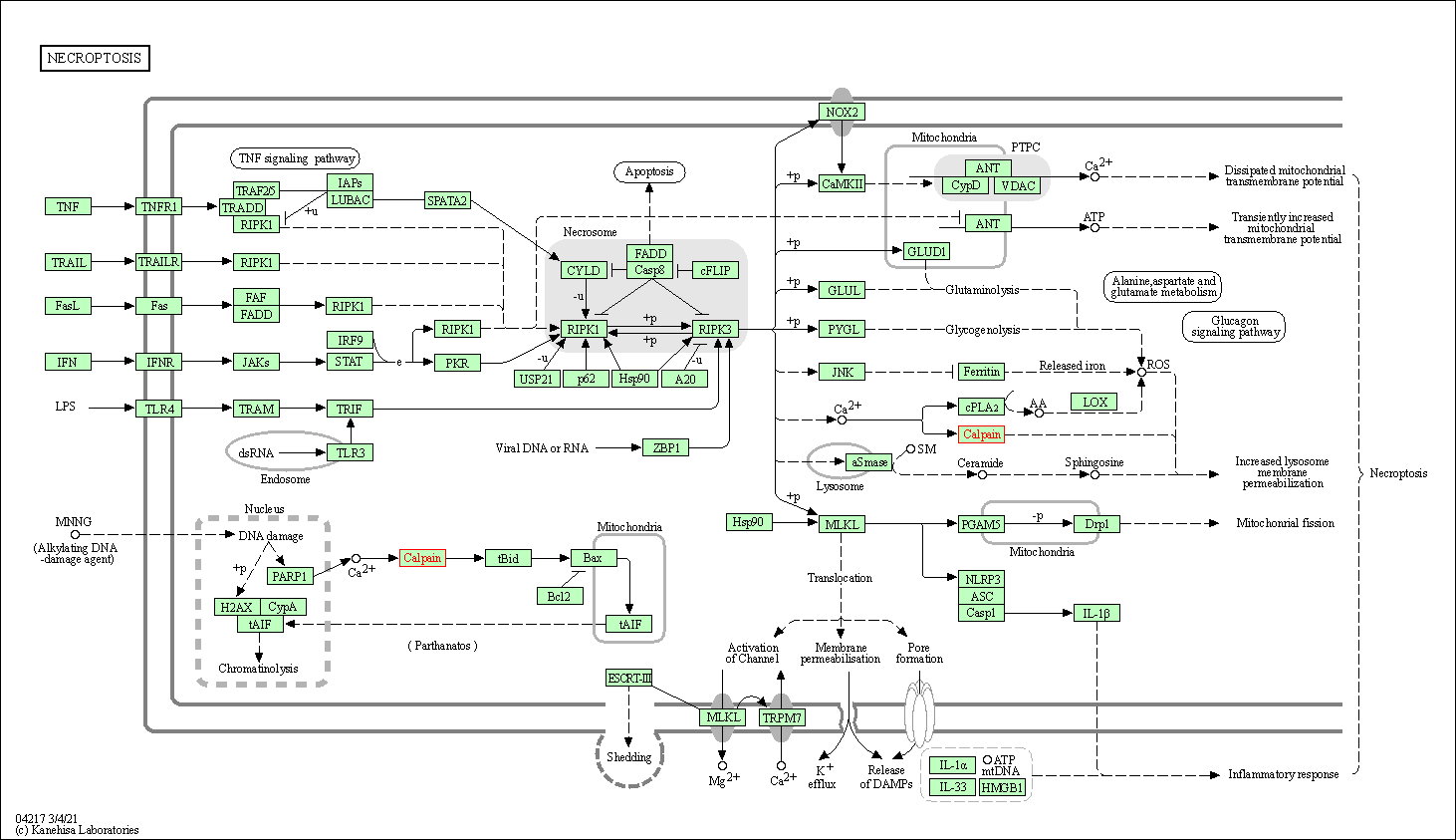
| Necroptosis | hsa04217 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
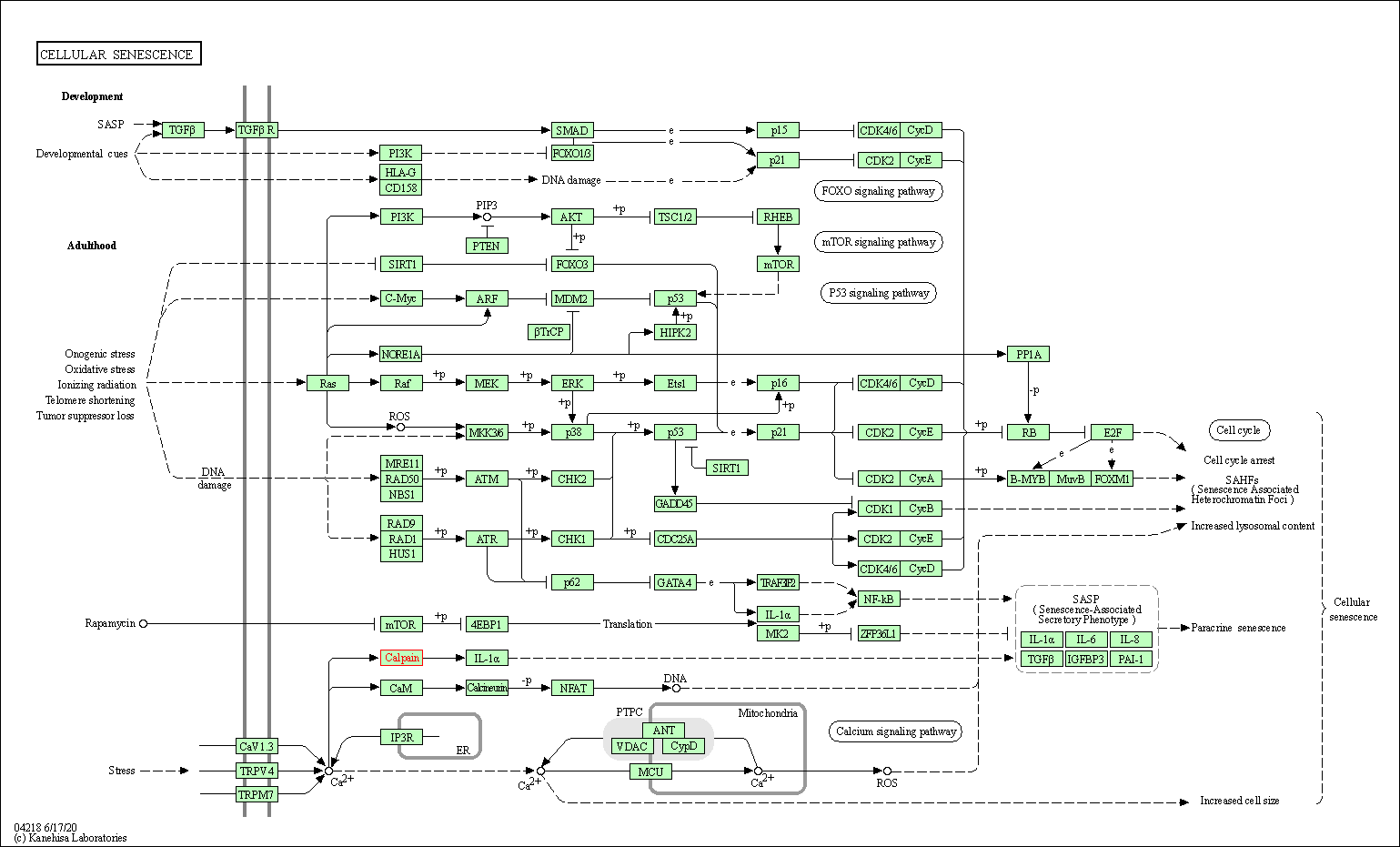
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 3.33E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.18E-01 | Radiality | 1.38E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 3.38E+01 | Topological coefficient | 2.40E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Protein processing in endoplasmic reticulum | |||||
| 2 | Apoptosis | |||||
| 3 | Alzheimer's disease | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Degradation of the extracellular matrix | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Focal Adhesion | |||||
| 2 | Integrated Pancreatic Cancer Pathway | |||||
| 3 | Alzheimers Disease | |||||
| 4 | Integrin-mediated Cell Adhesion | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 2 | ClinicalTrials.gov (NCT02220738) Study to Evaluate the Safety, Tolerability, and Pharmacokinetics of ABT-957 in Subjects With Mild-to-Moderate Alzheimer's Disease on Stable Doses of Acetylcholinesterase Inhibitors. U.S. National Institutes of Health. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 2338). | |||||
| REF 4 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 5 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800002441) | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800007674) | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800003692) | |||||
| REF 8 | An updated patent review of calpain inhibitors (2012 - 2014).Expert Opin Ther Pat. 2015 Jan;25(1):17-31. | |||||
| REF 9 | In silico identification and biochemical evaluation of novel inhibitors of NRH:quinone oxidoreductase 2 (NQO2). Bioorg Med Chem Lett. 2010 Dec 15;20(24):7331-6. | |||||
| REF 10 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2336). | |||||
| REF 11 | Synthesis of a small library of diketopiperazines as potential inhibitors of calpain. Bioorg Med Chem Lett. 2005 Jun 15;15(12):3034-8. | |||||
| REF 12 | Synthesis, biological evaluation and molecular modelling of N-heterocyclic dipeptide aldehydes as selective calpain inhibitors. Bioorg Med Chem. 2008 Jul 15;16(14):6911-23. | |||||
| REF 13 | Alpha-diketone and alpha-keto ester derivatives of N-protected amino acids and peptides as novel inhibitors of cysteine and serine proteinases. J Med Chem. 1990 Jan;33(1):11-3. | |||||
| REF 14 | Synthesis of cystamidin a (pyrrole-3-propanamide), a reported calpain inhibitor, Bioorg. Med. Chem. Lett. 6(13):1541-1542 (1996). | |||||
| REF 15 | Calpain in the pathophysiology of spinal cord injury: neuroprotection with calpain inhibitors. Brain Res Brain Res Rev. 2003 May;42(2):169-85. | |||||
| REF 16 | PEST sequences in the malaria parasite Plasmodium falciparum: a genomic study. Malar J. 2003 Jun 23;2:16. | |||||
| REF 17 | Peptidyl alpha-ketoamides with nucleobases, methylpiperazine, and dimethylaminoalkyl substituents as calpain inhibitors. J Med Chem. 2010 Sep 9;53(17):6326-36. | |||||
| REF 18 | Novel peptidyl alpha-keto amide inhibitors of calpains and other cysteine proteases. J Med Chem. 1996 Sep 27;39(20):4089-98. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

