Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T23606
(Former ID: TTDI03492)
|
|||||
| Target Name |
Protein kinase G2 (PRKG2)
|
|||||
| Synonyms |
cGMP-dependent protein kinase II; cGMP-dependent protein kinase 2; cGKII; cGK2; cGK 2; PRKGR2
Click to Show/Hide
|
|||||
| Gene Name |
PRKG2
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Crucial regulator of intestinal secretion and bone growth (By similarity). Phosphorylates and activates CFTR on the plasma membrane. Plays a key role in intestinal secretion by regulating cGMP-dependent translocation of CFTR in jejunum (By similarity). Acts downstream of NMDAR to activate the plasma membrane accumulation of GRIA1/GLUR1 in synapse and increase synaptic plasticity. Phosphorylates GRIA1/GLUR1 at Ser-863 (By similarity). Acts as regulator of gene expression and activator of the extracellular signal-regulated kinases MAPK3/ERK1 and MAPK1/ERK2 in mechanically stimulated osteoblasts. Under fluid shear stress, mediates ERK activation and subsequent induction of FOS, FOSL1/FRA1, FOSL2/FRA2 and FOSB that play a key role in the osteoblast anabolic response to mechanical stimulation (By similarity).
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.12
|
|||||
| Sequence |
MGNGSVKPKHSKHPDGHSGNLTTDALRNKVTELERELRRKDAEIQEREYHLKELREQLSK
QTVAIAELTEELQNKCIQLNKLQDVVHMQGGSPLQASPDKVPLEVHRKTSGLVSLHSRRG AKAGVSAEPTTRTYDLNKPPEFSFEKARVRKDSSEKKLITDALNKNQFLKRLDPQQIKDM VECMYGRNYQQGSYIIKQGEPGNHIFVLAEGRLEVFQGEKLLSSIPMWTTFGELAILYNC TRTASVKAITNVKTWALDREVFQNIMRRTAQARDEQYRNFLRSVSLLKNLPEDKLTKIID CLEVEYYDKGDYIIREGEEGSTFFILAKGKVKVTQSTEGHDQPQLIKTLQKGEYFGEKAL ISDDVRSANIIAEENDVACLVIDRETFNQTVGTFEELQKYLEGYVANLNRDDEKRHAKRS MSNWKLSKALSLEMIQLKEKVARFSSSSPFQNLEIIATLGVGGFGRVELVKVKNENVAFA MKCIRKKHIVDTKQQEHVYSEKRILEELCSPFIVKLYRTFKDNKYVYMLLEACLGGELWS ILRDRGSFDEPTSKFCVACVTEAFDYLHRLGIIYRDLKPENLILDAEGYLKLVDFGFAKK IGSGQKTWTFCGTPEYVAPEVILNKGHDFSVDFWSLGILVYELLTGNPPFSGVDQMMTYN LILKGIEKMDFPRKITRRPEDLIRRLCRQNPTERLGNLKNGINDIKKHRWLNGFNWEGLK ARSLPSPLQRELKGPIDHSYFDKYPPEKGMPPDELSGWDKDF Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Cyclic Guanosine Monophosphate | Ligand Info | |||||
| Structure Description | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with cGMP | PDB:5BV6 | ||||
| Method | X-ray diffraction | Resolution | 1.94 Å | Mutation | No | [2] |
| PDB Sequence |
TAQARDEQYR
278 NFLRSVSLLK288 NLPEDKLTKI298 IDCLEVEYYD308 KGDYIIREGE318 EGSTFFILAK 328 GKVKVTQSTE338 GHDQPQLIKT348 LQKGEYFGEK358 ALISDDVRSA368 NIIAEENDVA 378 CLVIDRETFN388 QTVGTFEELQ398 KYLEGYVANL408 NRDDEKRHAK418 |
|||||
|
|
ILE314
3.906
VAL333
3.826
GLN335
3.535
ILE346
4.103
LYS347
3.183
TYR354
4.584
PHE355
3.280
GLY356
2.968
GLU357
2.647
LYS358
3.295
ALA359
3.015
|
|||||
| Ligand Name: Cyclic Guanosine Monophosphate | Ligand Info | |||||
| Structure Description | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cGMP | PDB:5C8W | ||||
| Method | X-ray diffraction | Resolution | 1.80 Å | Mutation | No | [2] |
| PDB Sequence |
RKDSSEKKLI
159 TDALNKNQFL169 KRLDPQQIKD179 MVECMYGRNY189 QQGSYIIKQG199 EPGNHIFVLA 209 EGRLEVFQGE219 KLLSSIPMWT229 TFGELAILYN239 CTRTASVKAI249 TNVKTWALDR 259 EVFQNIMRRT269
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
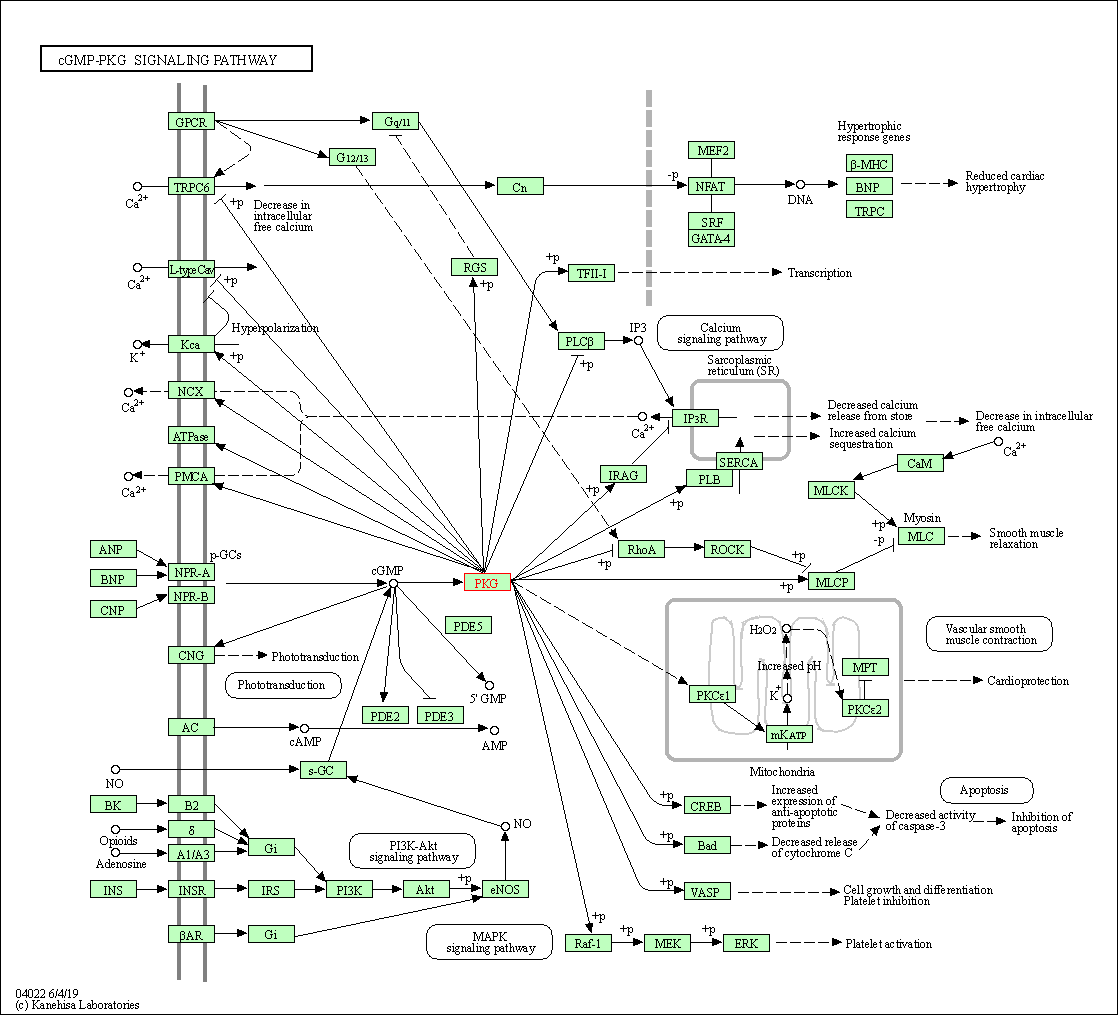
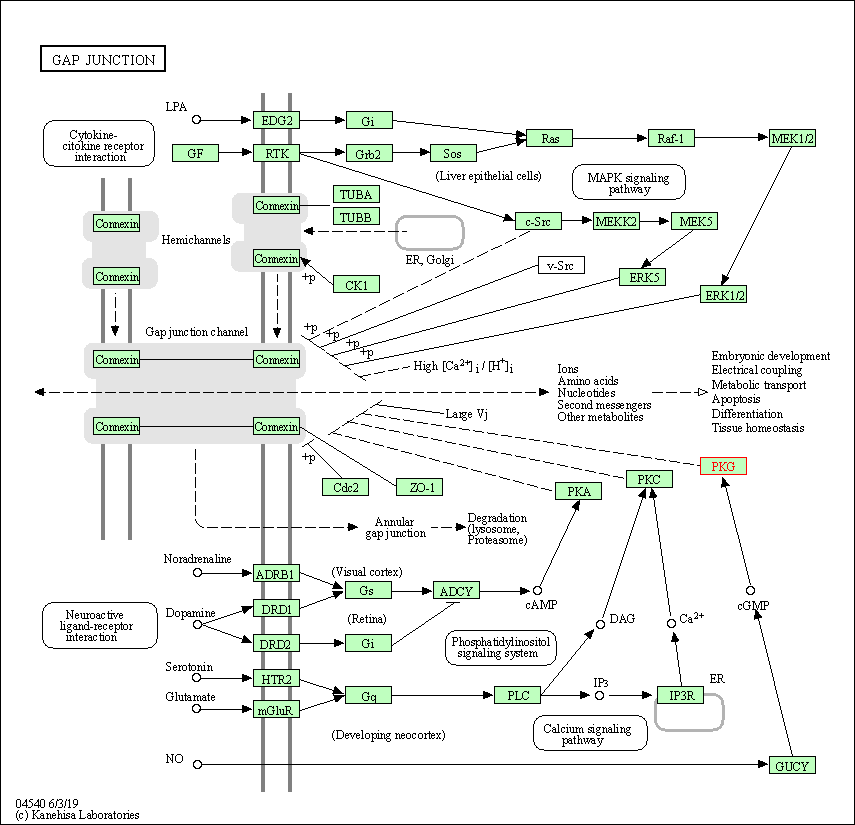
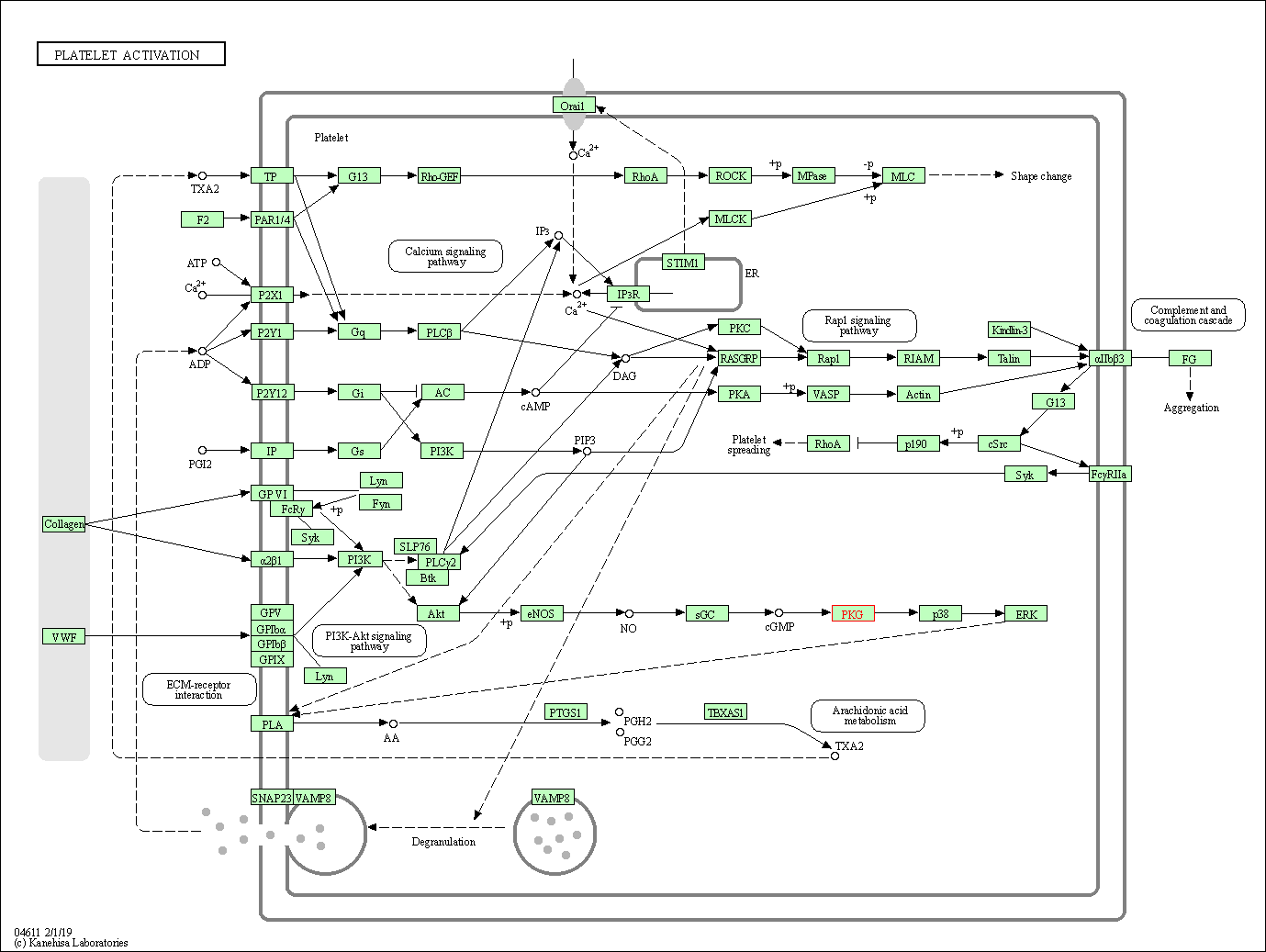
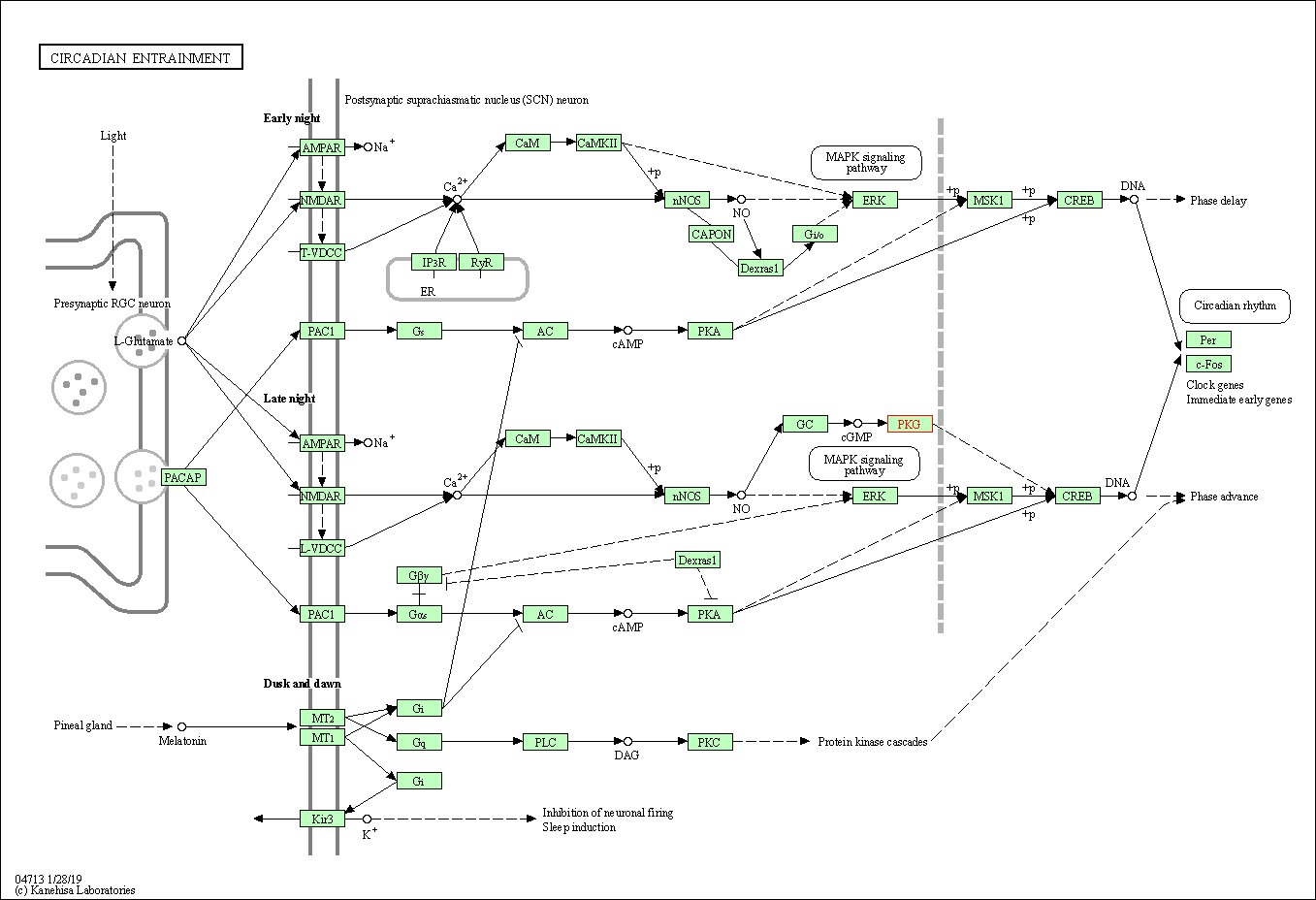
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Gap junction | hsa04540 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Circadian entrainment | hsa04713 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
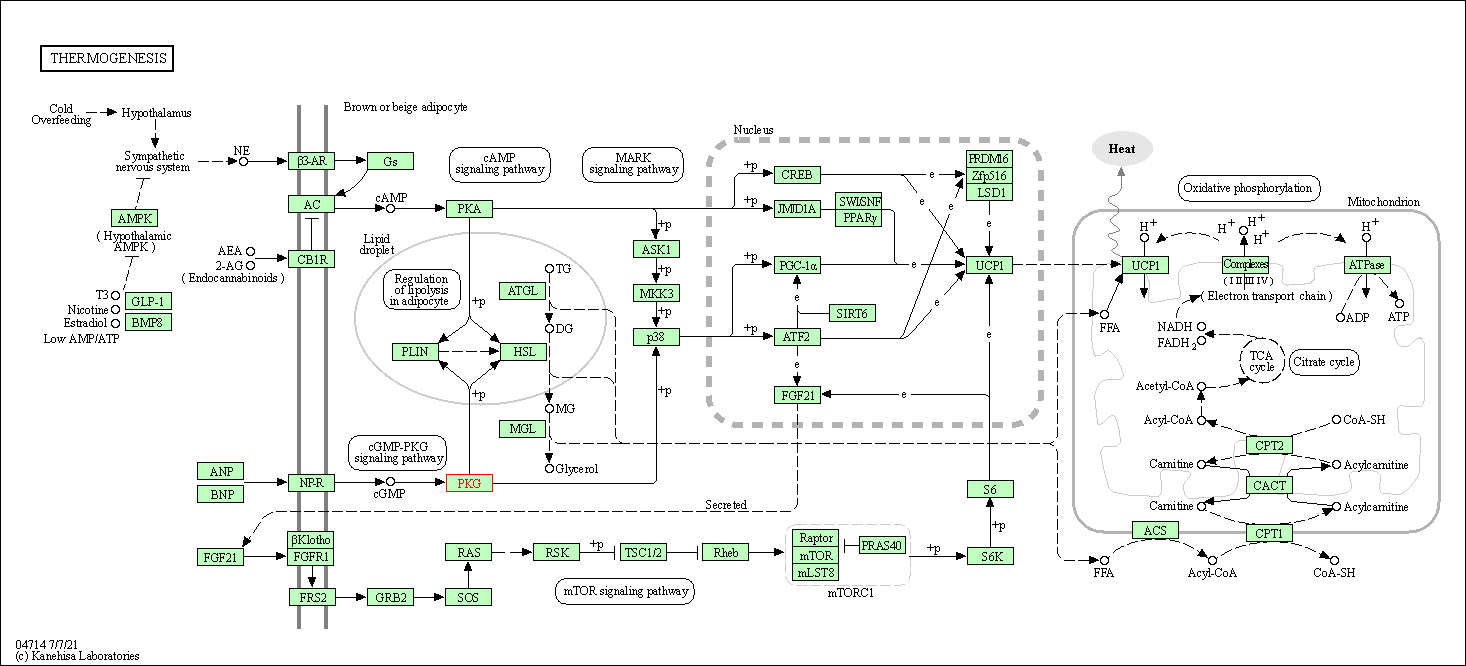
| Thermogenesis | hsa04714 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
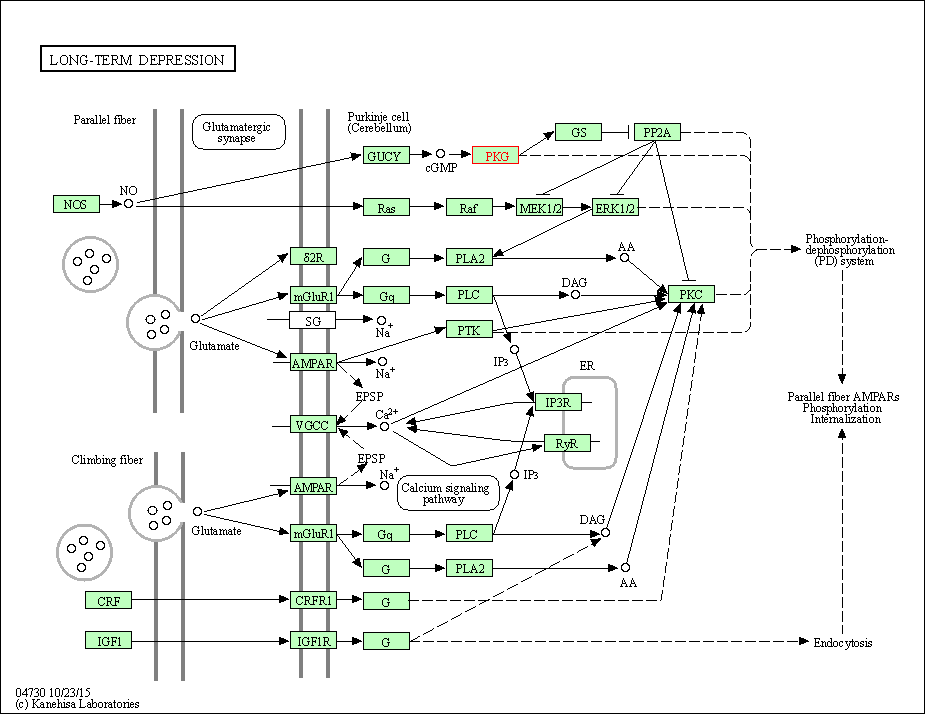
| Long-term depression | hsa04730 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
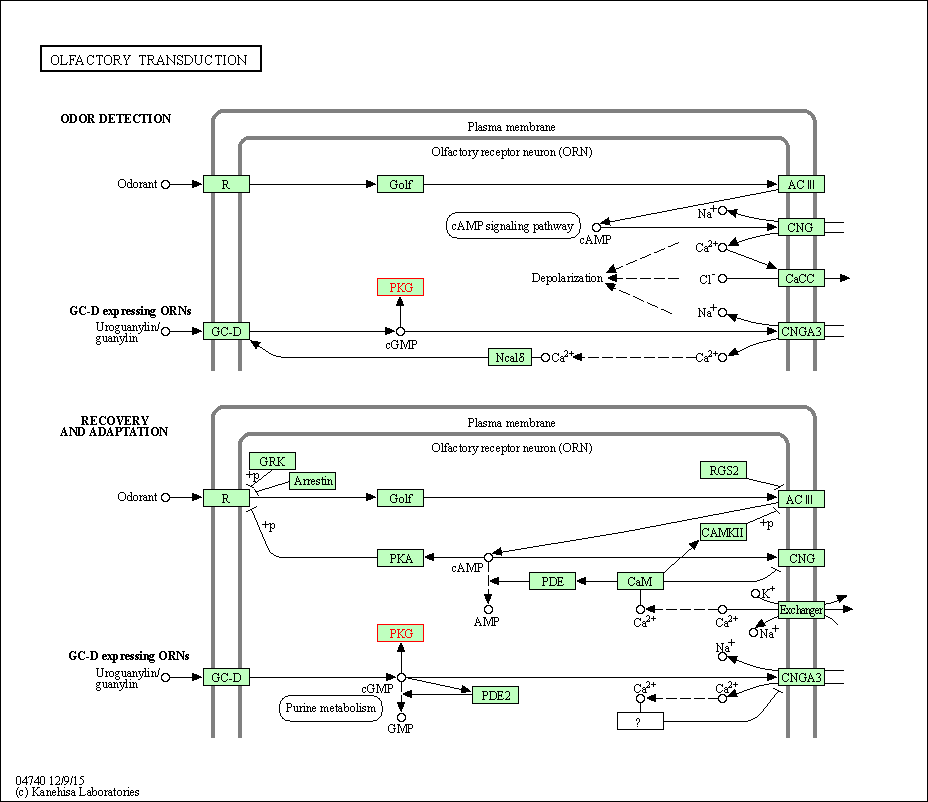
| Olfactory transduction | hsa04740 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
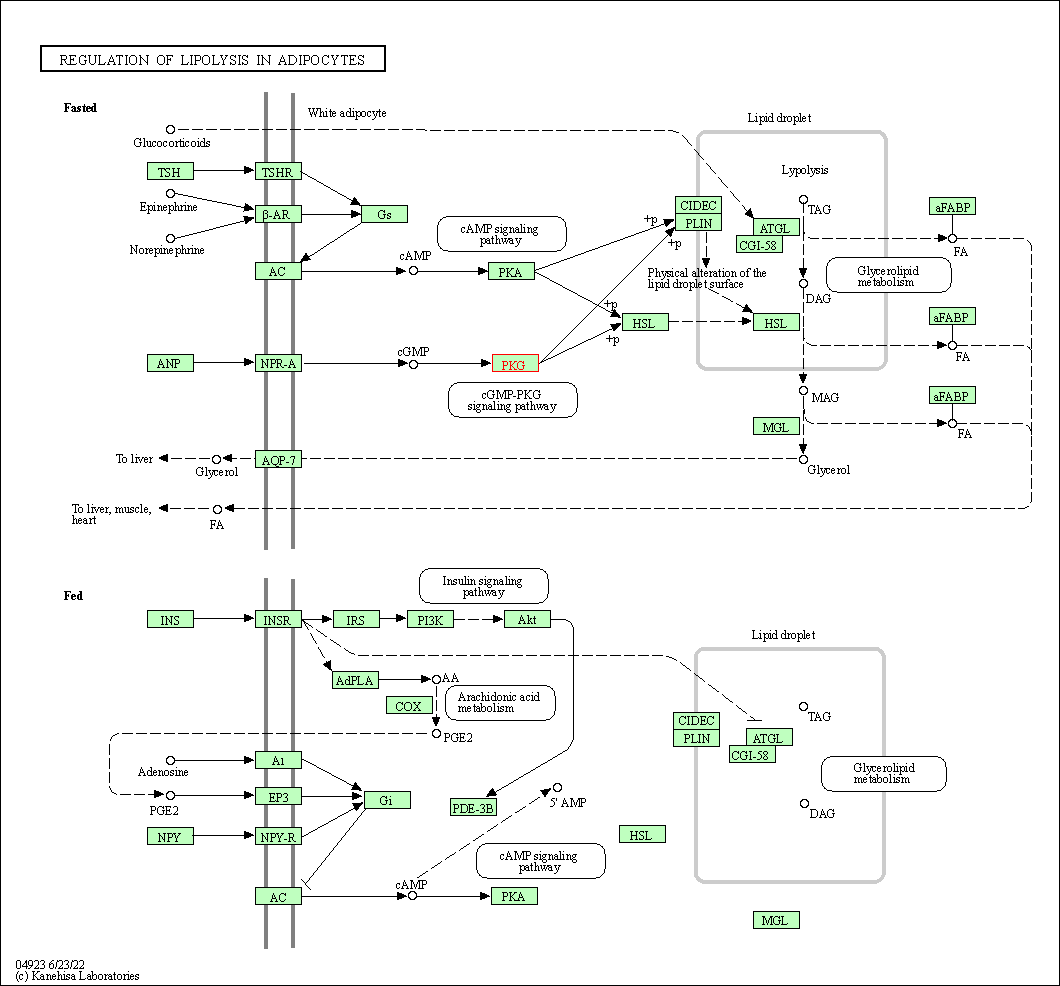
| Regulation of lipolysis in adipocytes | hsa04923 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
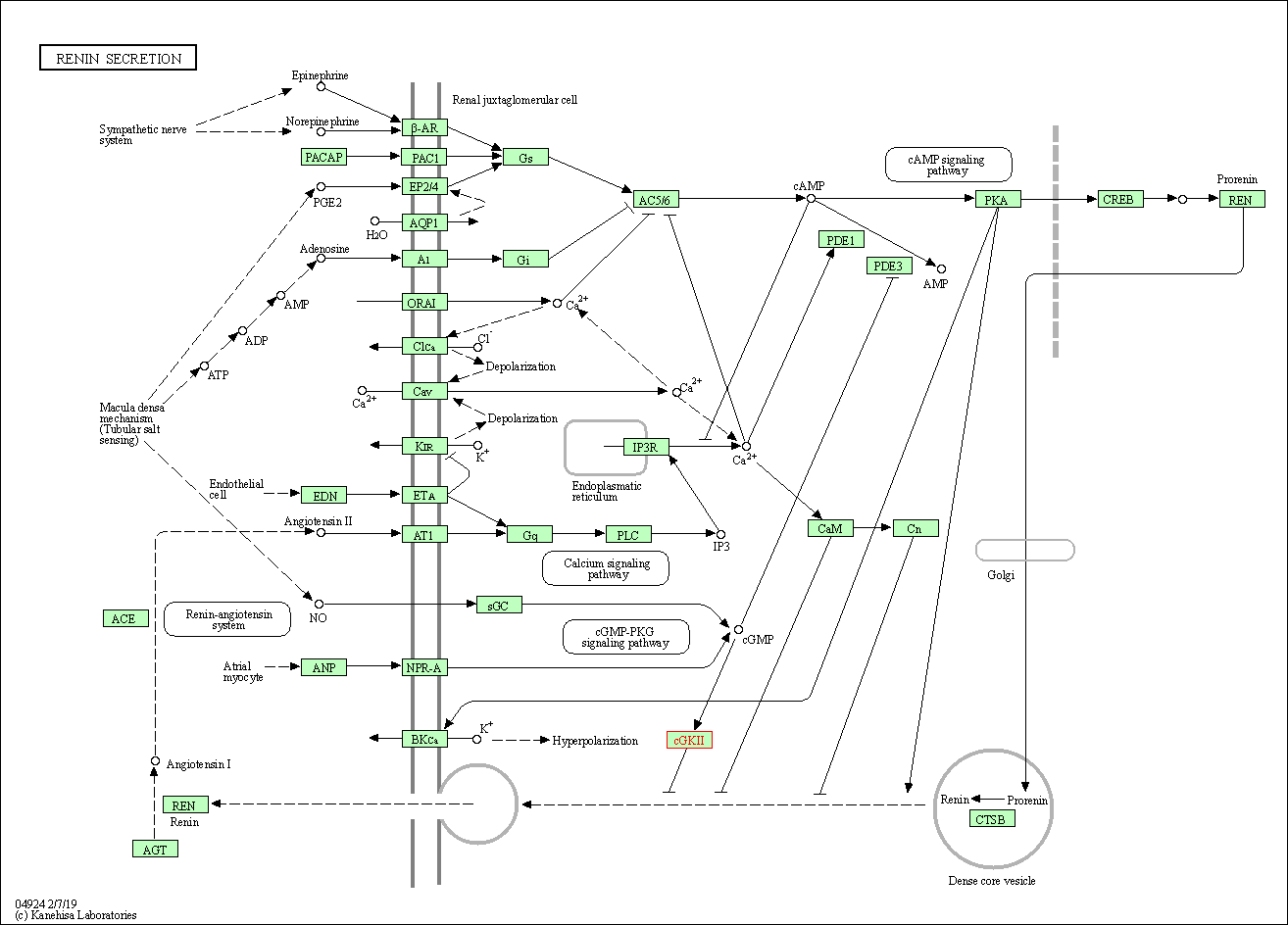
| Renin secretion | hsa04924 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
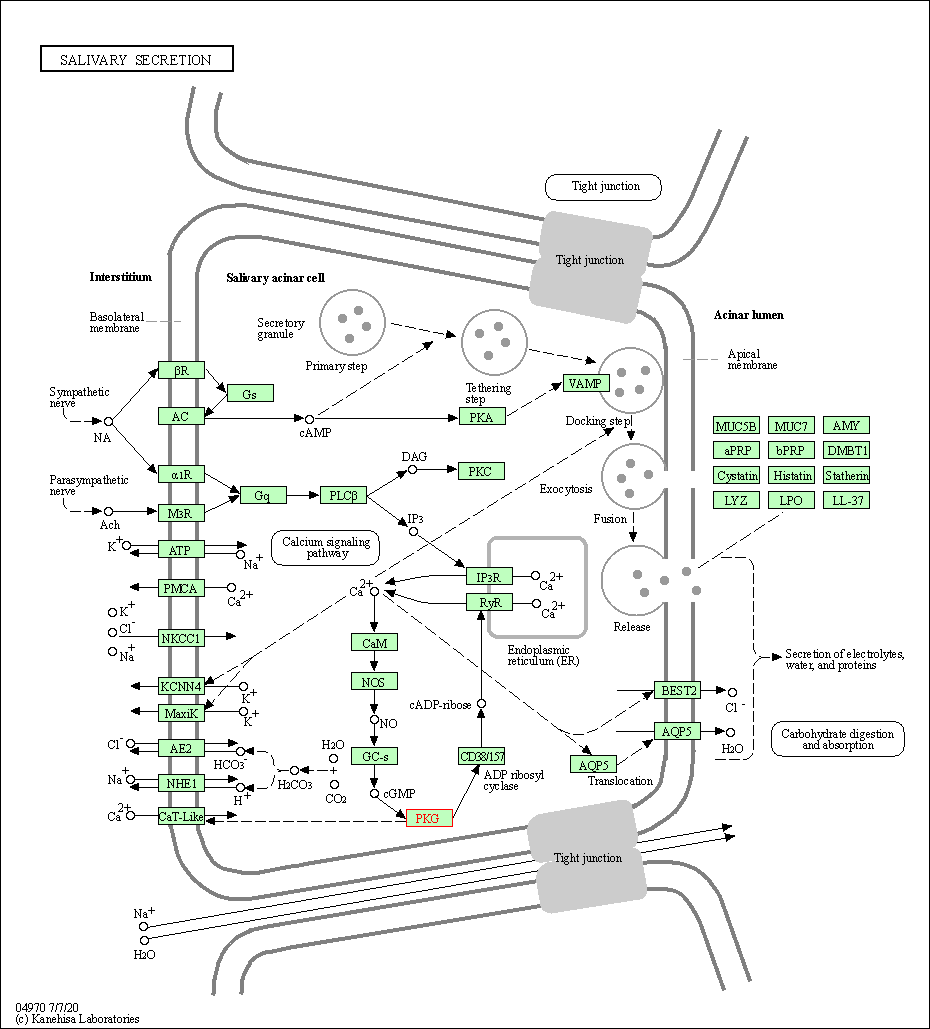
| Salivary secretion | hsa04970 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.58E-01 | Radiality | 1.23E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 3.00E+00 | Topological coefficient | 1.00E+00 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Substituted 2H-isoquinolin-1-ones as potent Rho-kinase inhibitors: part 3, aryl substituted pyrrolidines. Bioorg Med Chem Lett. 2010 Jun 15;20(12):3746-9. | |||||
| REF 2 | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II. J Biol Chem. 2016 Mar 11;291(11):5623-5633. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

