Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T26617
(Former ID: TTDR00154)
|
|||||
| Target Name |
Lactase-phlorizin hydrolase (LCT)
|
|||||
| Synonyms |
Lactase-glycosylceramidase; LCT; Glycosyl-hydrolase
Click to Show/Hide
|
|||||
| Gene Name |
LCT
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
LPH splits lactose in the small intestine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Glycosylase
|
|||||
| UniProt ID | ||||||
| Sequence |
MELSWHVVFIALLSFSCWGSDWESDRNFISTAGPLTNDLLHNLSGLLGDQSSNFVAGDKD
MYVCHQPLPTFLPEYFSSLHASQITHYKVFLSWAQLLPAGSTQNPDEKTVQCYRRLLKAL KTARLQPMVILHHQTLPASTLRRTEAFADLFADYATFAFHSFGDLVGIWFTFSDLEEVIK ELPHQESRASQLQTLSDAHRKAYEIYHESYAFQGGKLSVVLRAEDIPELLLEPPISALAQ DTVDFLSLDLSYECQNEASLRQKLSKLQTIEPKVKVFIFNLKLPDCPSTMKNPASLLFSL FEAINKDQVLTIGFDINEFLSCSSSSKKSMSCSLTGSLALQPDQQQDHETTDSSPASAYQ RIWEAFANQSRAERDAFLQDTFPEGFLWGASTGAFNVEGGWAEGGRGVSIWDPRRPLNTT EGQATLEVASDSYHKVASDVALLCGLRAQVYKFSISWSRIFPMGHGSSPSLPGVAYYNKL IDRLQDAGIEPMATLFHWDLPQALQDHGGWQNESVVDAFLDYAAFCFSTFGDRVKLWVTF HEPWVMSYAGYGTGQHPPGISDPGVASFKVAHLVLKAHARTWHHYNSHHRPQQQGHVGIV LNSDWAEPLSPERPEDLRASERFLHFMLGWFAHPVFVDGDYPATLRTQIQQMNRQCSHPV AQLPEFTEAEKQLLKGSADFLGLSHYTSRLISNAPQNTCIPSYDTIGGFSQHVNHVWPQT SSSWIRVVPWGIRRLLQFVSLEYTRGKVPIYLAGNGMPIGESENLFDDSLRVDYFNQYIN EVLKAIKEDSVDVRSYIARSLIDGFEGPSGYSQRFGLHHVNFSDSSKSRTPRKSAYFFTS IIEKNGFLTKGAKRLLPPNTVNLPSKVRAFTFPSEVPSKAKVVWEKFSSQPKFERDLFYH GTFRDDFLWGVSSSAYQIEGAWDADGKGPSIWDNFTHTPGSNVKDNATGDIACDSYHQLD ADLNMLRALKVKAYRFSISWSRIFPTGRNSSINSHGVDYYNRLINGLVASNIFPMVTLFH WDLPQALQDIGGWENPALIDLFDSYADFCFQTFGDRVKFWMTFNEPMYLAWLGYGSGEFP PGVKDPGWAPYRIAHAVIKAHARVYHTYDEKYRQEQKGVISLSLSTHWAEPKSPGVPRDV EAADRMLQFSLGWFAHPIFRNGDYPDTMKWKVGNRSELQHLATSRLPSFTEEEKRFIRAT ADVFCLNTYYSRIVQHKTPRLNPPSYEDDQEMAEEEDPSWPSTAMNRAAPWGTRRLLNWI KEEYGDIPIYITENGVGLTNPNTEDTDRIFYHKTYINEALKAYRLDGIDLRGYVAWSLMD NFEWLNGYTVKFGLYHVDFNNTNRPRTARASARYYTEVITNNGMPLAREDEFLYGRFPEG FIWSAASAAYQIEGAWRADGKGLSIWDTFSHTPLRVENDAIGDVACDSYHKIAEDLVTLQ NLGVSHYRFSISWSRILPDGTTRYINEAGLNYYVRLIDTLLAASIQPQVTIYHWDLPQTL QDVGGWENETIVQRFKEYADVLFQRLGDKVKFWITLNEPFVIAYQGYGYGTAAPGVSNRP GTAPYIVGHNLIKAHAEAWHLYNDVYRASQGGVISITISSDWAEPRDPSNQEDVEAARRY VQFMGGWFAHPIFKNGDYNEVMKTRIRDRSLAAGLNKSRLPEFTESEKRRINGTYDFFGF NHYTTVLAYNLNYATAISSFDADRGVASIADRSWPDSGSFWLKMTPFGFRRILNWLKEEY NDPPIYVTENGVSQREETDLNDTARIYYLRTYINEALKAVQDKVDLRGYTVWSAMDNFEW ATGFSERFGLHFVNYSDPSLPRIPKASAKFYASVVRCNGFPDPATGPHACLHQPDAGPTI SPVRQEEVQFLGLMLGTTEAQTALYVLFSLVLLGVCGLAFLSYKYCKRSKQGKTQRSQQE LSPVSSF Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T64DYJ | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
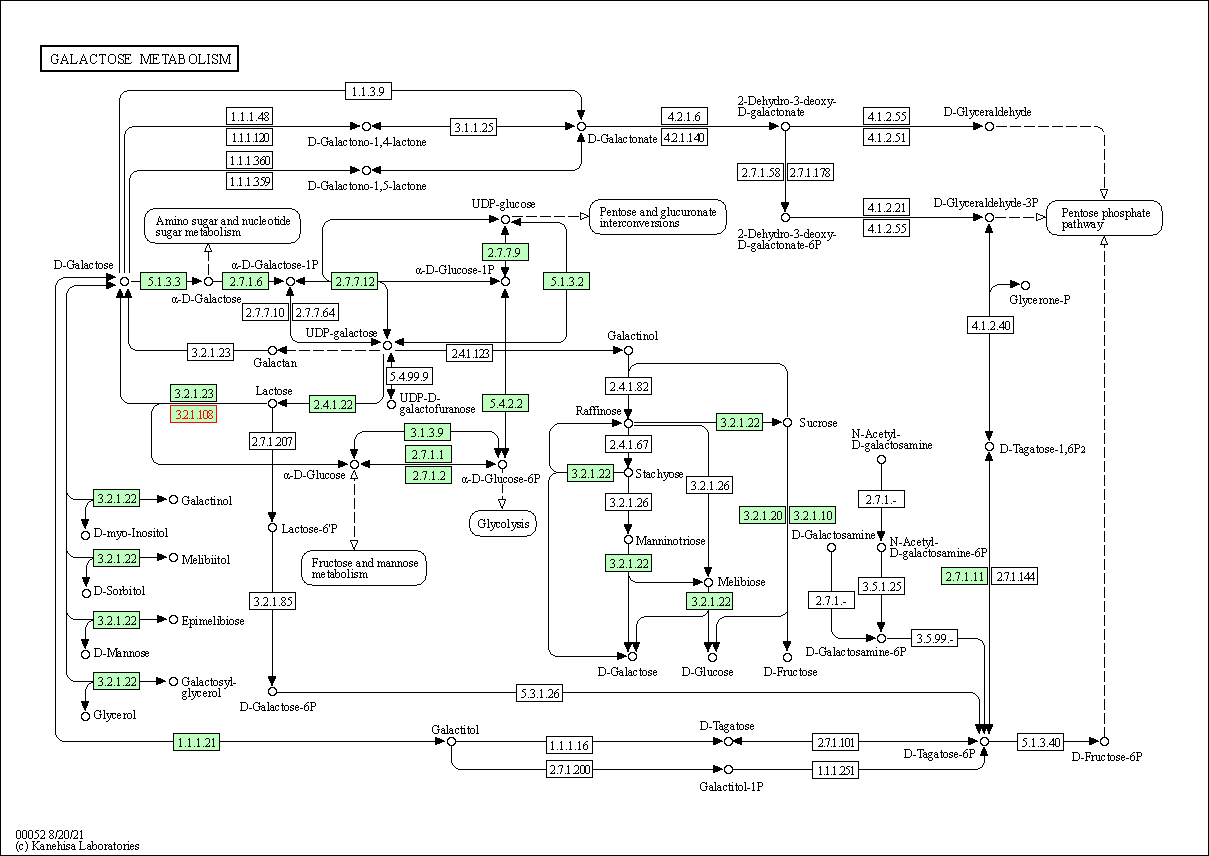
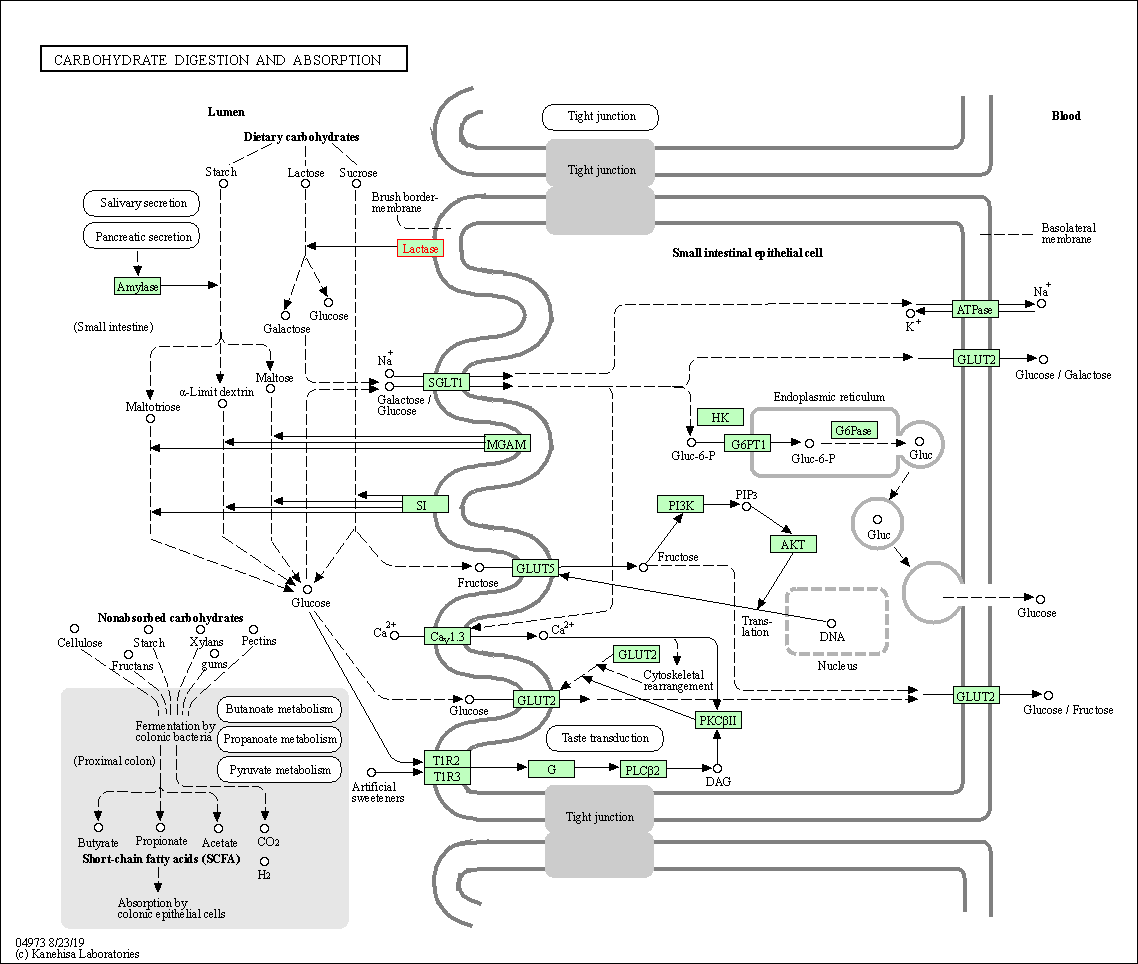
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Galactose metabolism | hsa00052 | Affiliated Target |

|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Carbohydrate digestion and absorption | hsa04973 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Galactose metabolism | |||||
| 2 | Metabolic pathways | |||||
| 3 | Carbohydrate digestion and absorption | |||||
| Pathwhiz Pathway | [+] 2 Pathwhiz Pathways | + | ||||
| 1 | Lactose Degradation | |||||
| 2 | Galactose Metabolism | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Metabolism of carbohydrates | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 2 | 2,5-Dideoxy-2,5-imino-d-altritol as a new class of pharmacological chaperone for Fabry disease. Bioorg Med Chem. 2010 Jun 1;18(11):3790-4. | |||||
| REF 3 | Nitrogen-containing furanose and pyranose analogues from Hyacinthus orientalis. J Nat Prod. 1998 May;61(5):625-8. | |||||
| REF 4 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 5 | New polyhydroxylated pyrrolidine, piperidine, and pyrrolizidine alkaloids from Scilla sibirica. J Nat Prod. 2002 Dec;65(12):1875-81. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

