Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T30163
(Former ID: TTDI03192)
|
|||||
| Target Name |
Excitatory amino acid transporter 4 (SLC1A6)
|
|||||
| Synonyms |
Solute carrier family 1 member 6; Sodium-dependent glutamate/aspartate transporter; EAAT4
Click to Show/Hide
|
|||||
| Gene Name |
SLC1A6
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Sodium-dependent, high-affinity amino acid transporter that mediates the uptake of L-glutamate and also L-aspartate and D-aspartate. Functions as a symporter that transports one amino acid molecule together with two or three Na(+) ions and one proton, in parallel with the counter-transport of one K(+) ion. Mediates Cl(-) flux that is not coupled to amino acid transport; this avoids the accumulation of negative charges due to aspartate and Na(+) symport (By similarity). Plays a redundant role in the rapid removal of released glutamate from the synaptic cleft, which is essential for terminating the postsynaptic action of glutamate (Probable).
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MSSHGNSLFLRESGQRLGRVGWLQRLQESLQQRALRTRLRLQTMTLEHVLRFLRRNAFIL
LTVSAVVIGVSLAFALRPYQLTYRQIKYFSFPGELLMRMLQMLVLPLIVSSLVTGMASLD NKATGRMGMRAAVYYMVTTIIAVFIGILMVTIIHPGKGSKEGLHREGRIETIPTADAFMD LIRNMFPPNLVEACFKQFKTQYSTRVVTRTMVRTENGSEPGASMPPPFSVENGTSFLENV TRALGTLQEMLSFEETVPVPGSANGINALGLVVFSVAFGLVIGGMKHKGRVLRDFFDSLN EAIMRLVGIIIWYAPVGILFLIAGKILEMEDMAVLGGQLGMYTLTVIVGLFLHAGIVLPL IYFLVTHRNPFPFIGGMLQALITAMGTSSSSATLPITFRCLEEGLGVDRRITRFVLPVGA TVNMDGTALYEALAAIFIAQVNNYELNLGQITTISITATAASVGAAGIPQAGLVTMVIVL TSVGLPTEDITLIIAVDWFLDRLRTMTNVLGDSIGAAVIEHLSQRELELQEAELTLPSLG KPYKSLMAQEKGASRGRGGNESAM Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T17B47 | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
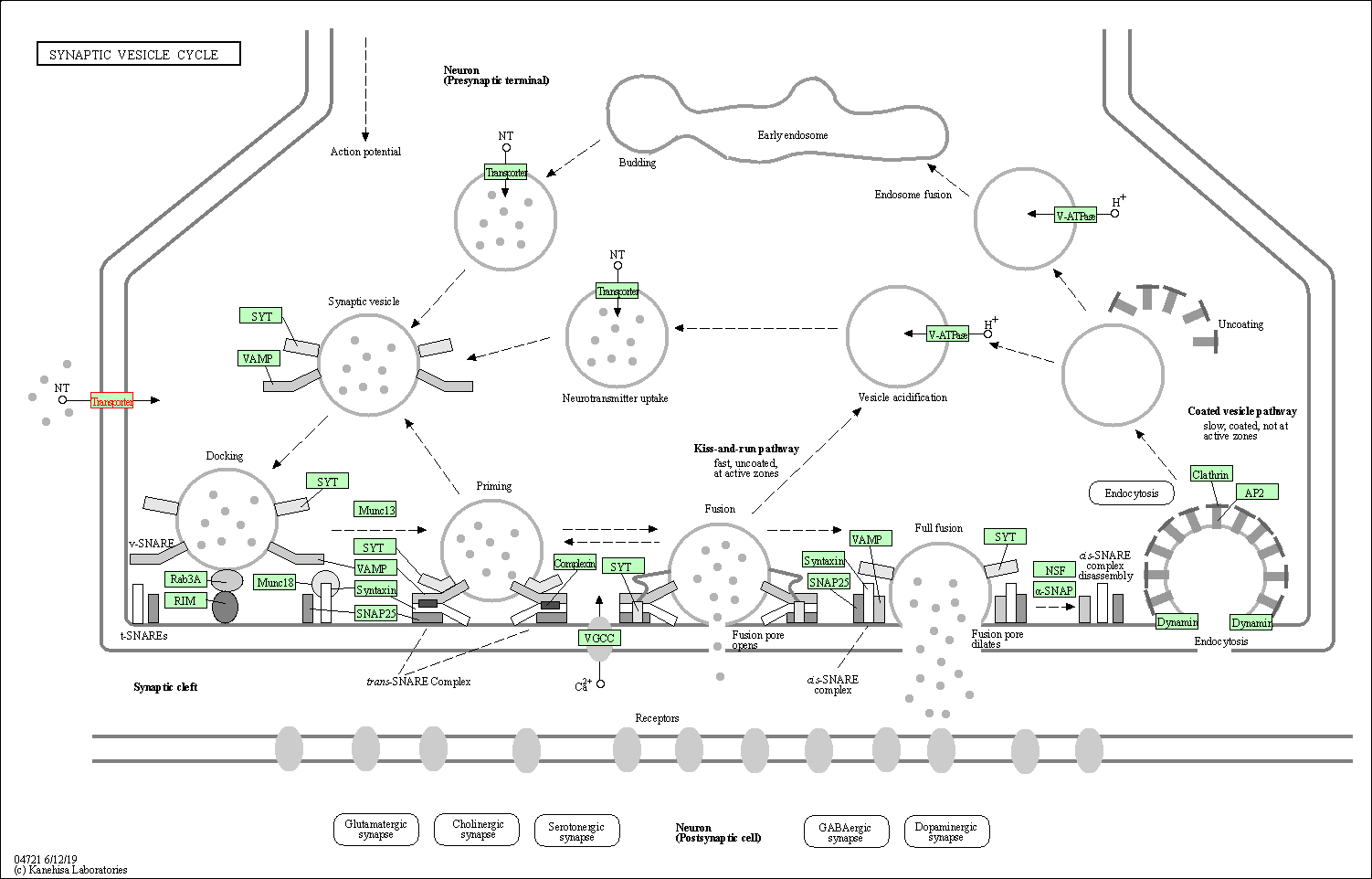
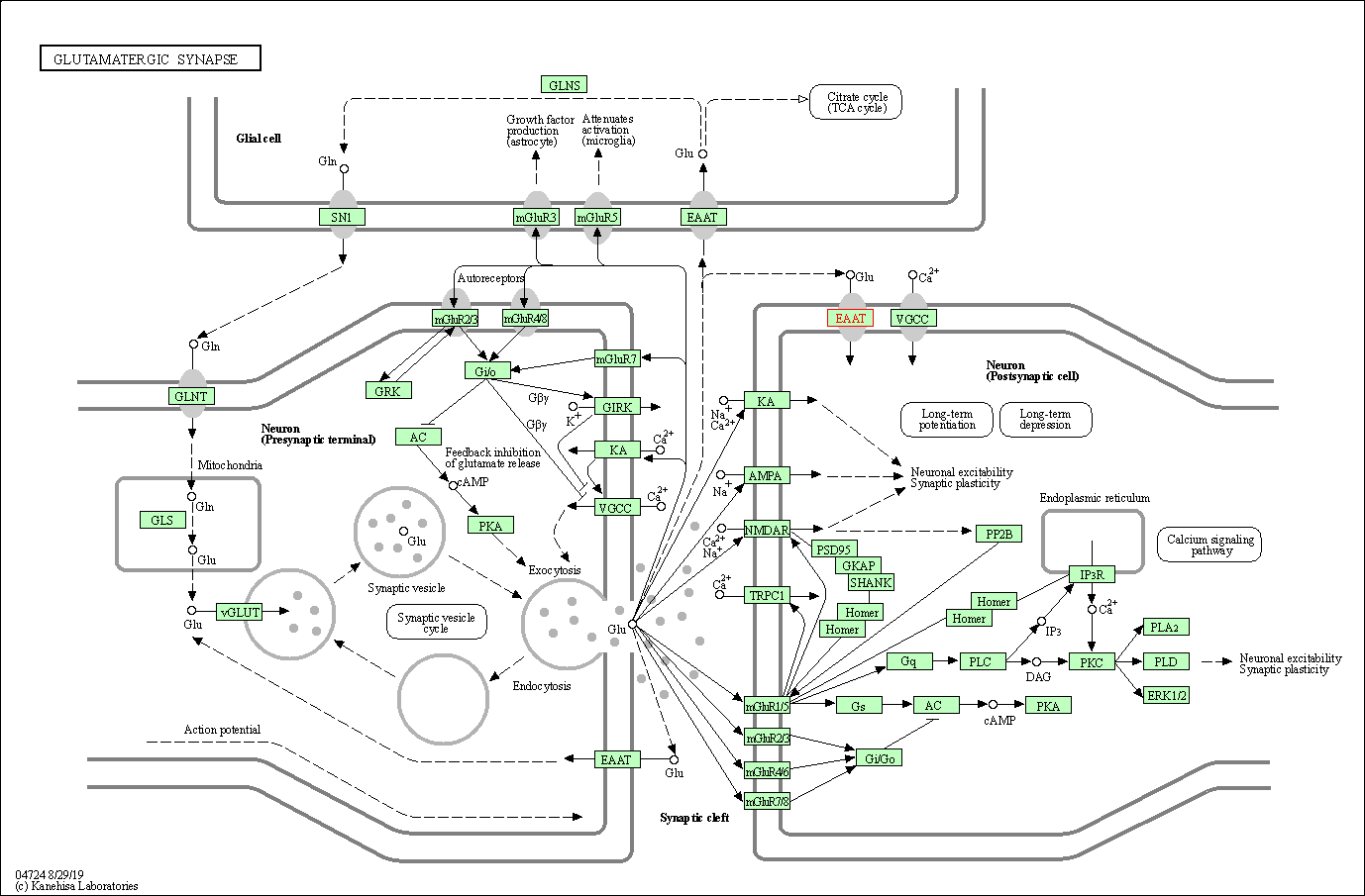
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Synaptic vesicle cycle | hsa04721 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Glutamatergic synapse | hsa04724 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Pharmacological characterization of threo-3-methylglutamic acid with excitatory amino acid transporters in native and recombinant systems. J Neurochem. 2001 Apr;77(2):550-7. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 871). | |||||
| REF 3 | Effects of threo-beta-hydroxyaspartate derivatives on excitatory amino acid transporters (EAAT4 and EAAT5). J Neurochem. 2001 Oct;79(2):297-302. | |||||
| REF 4 | Characterization of the tritium-labeled analog of L-threo-beta-benzyloxyaspartate binding to glutamate transporters. Mol Pharmacol. 2007 Jan;71(1):294-302. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

