Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T42247
(Former ID: TTDI03049)
|
|||||
| Target Name |
Melanoma inhibitor of apoptosis protein (ML-IAP)
|
|||||
| Synonyms |
UNQ5800/PRO19607/PRO21344; RNF50; RING-type E3 ubiquitin transferase BIRC7; RING finger protein 50; MLIAP; Livin; Kidney inhibitor of apoptosis protein; KIAP; Baculoviral IAP repeat-containing protein 7
Click to Show/Hide
|
|||||
| Gene Name |
BIRC7
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Function |
Its anti-apoptotic activity is mediated through the inhibition of CASP3, CASP7 and CASP9, as well as by its E3 ubiquitin-protein ligase activity. As it is a weak caspase inhibitor, its anti-apoptotic activity is thought to be due to its ability to ubiquitinate DIABLO/SMAC targeting it for degradation thereby promoting cell survival. May contribute to caspase inhibition, by blocking the ability of DIABLO/SMAC to disrupt XIAP/BIRC4-caspase interactions. Protects against apoptosis induced by TNF or by chemical agents such as adriamycin, etoposide or staurosporine. Suppression of apoptosis is mediated by activation of MAPK8/JNK1, and possibly also of MAPK9/JNK2. This activation depends on TAB1 and NR2C2/TAK1. In vitro, inhibits CASP3 and proteolytic activation of pro-CASP9. Isoform 1 blocks staurosporine-induced apoptosis. Isoform 2 blocks etoposide-induced apoptosis. Isoform 2 protects against natural killer (NK) cell killing whereas isoform 1 augments killing. Apoptotic regulator capable of exerting proapoptotic and anti-apoptotic activities and plays crucial roles in apoptosis, cell proliferation, and cell cycle control.
Click to Show/Hide
|
|||||
| BioChemical Class |
Acyltransferase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.3.2.27
|
|||||
| Sequence |
MGPKDSAKCLHRGPQPSHWAAGDGPTQERCGPRSLGSPVLGLDTCRAWDHVDGQILGQLR
PLTEEEEEEGAGATLSRGPAFPGMGSEELRLASFYDWPLTAEVPPELLAAAGFFHTGHQD KVRCFFCYGGLQSWKRGDDPWTEHAKWFPSCQFLLRSKGRDFVHSVQETHSQLLGSWDPW EEPEDAAPVAPSVPASGYPELPTPRREVQSESAQEPGGVSPAEAQRAWWVLEPPGARDVE AQLRRLQEERTCKVCLDRAVSIVFVPCGHLVCAECAPGLQLCPICRAPVRSRVRTFLS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T16S8T | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: N-{(1s)-1-Cyclohexyl-2-Oxo-2-[(2s)-2-(4-Phenyl-1,3-Benzothiazol-2-Yl)pyrrolidin-1-Yl]ethyl}-N~2~-Methyl-L-Alaninamide | Ligand Info | |||||
| Structure Description | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | PDB:3GTA | ||||
| Method | X-ray diffraction | Resolution | 1.70 Å | Mutation | Yes | [2] |
| PDB Sequence |
GPAFPGMGSE
87 ELRLASFYDW97 PLTAEVPPEL107 LAAAGFFHTG117 HQDKVRCFFC127 YGGLQSWKRG 137 DDPWTEHAKW147 FPGCQFLLRS157 KGQEYINNIH167
|
|||||
|
|
||||||
| Ligand Name: N-{(1s)-1-Cyclohexyl-2-[(2s)-2-(4-Naphthalen-1-Yl-1,3-Thiazol-2-Yl)pyrrolidin-1-Yl]-2-Oxoethyl}-N~2~-Methyl-L-Alaninamide | Ligand Info | |||||
| Structure Description | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | PDB:3GT9 | ||||
| Method | X-ray diffraction | Resolution | 1.70 Å | Mutation | Yes | [2] |
| PDB Sequence |
GPAFPGMGSE
87 ELRLASFYDW97 PLTAEVPPEL107 LAAAGFFHTG117 HQDKVRCFFC127 YGGLQSWKRG 137 DDPWTEHAKW147 FPGCQFLLRS157 KGQEYINNIH167
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
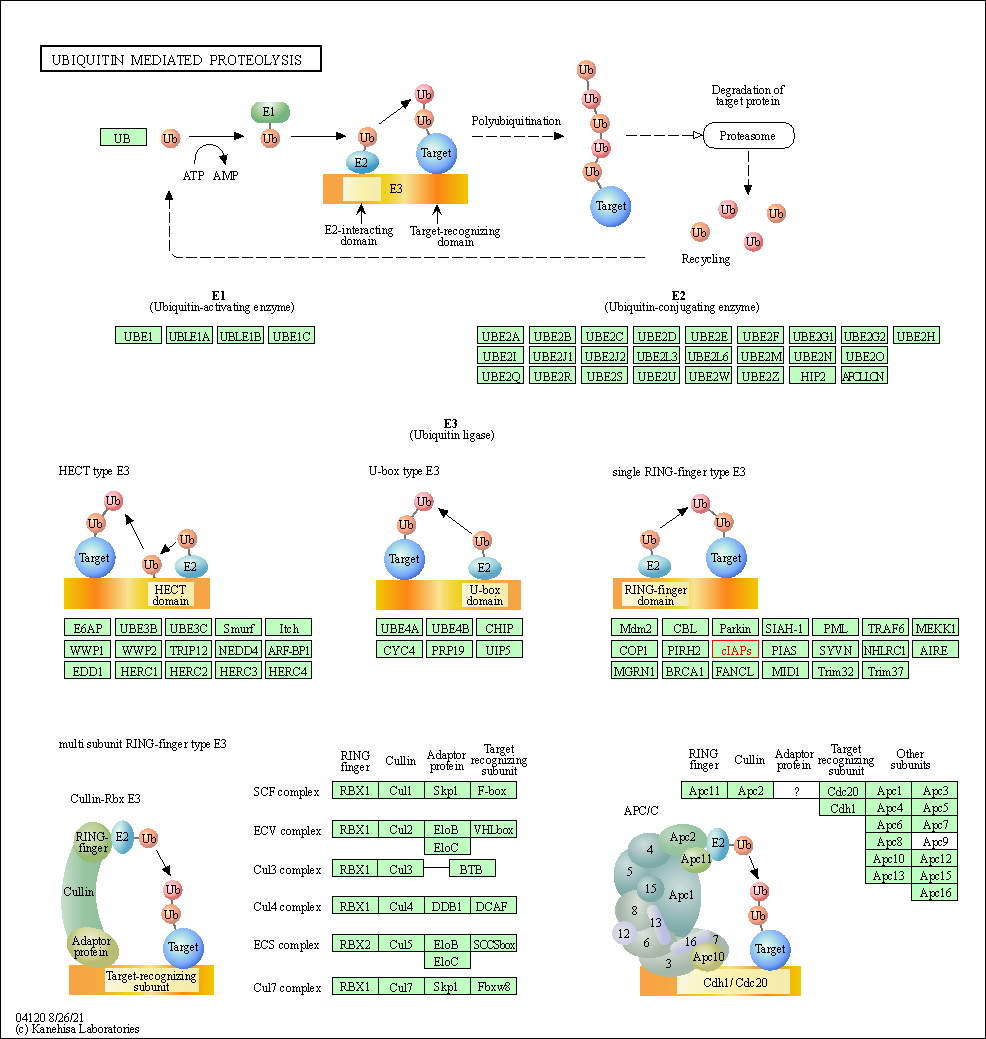
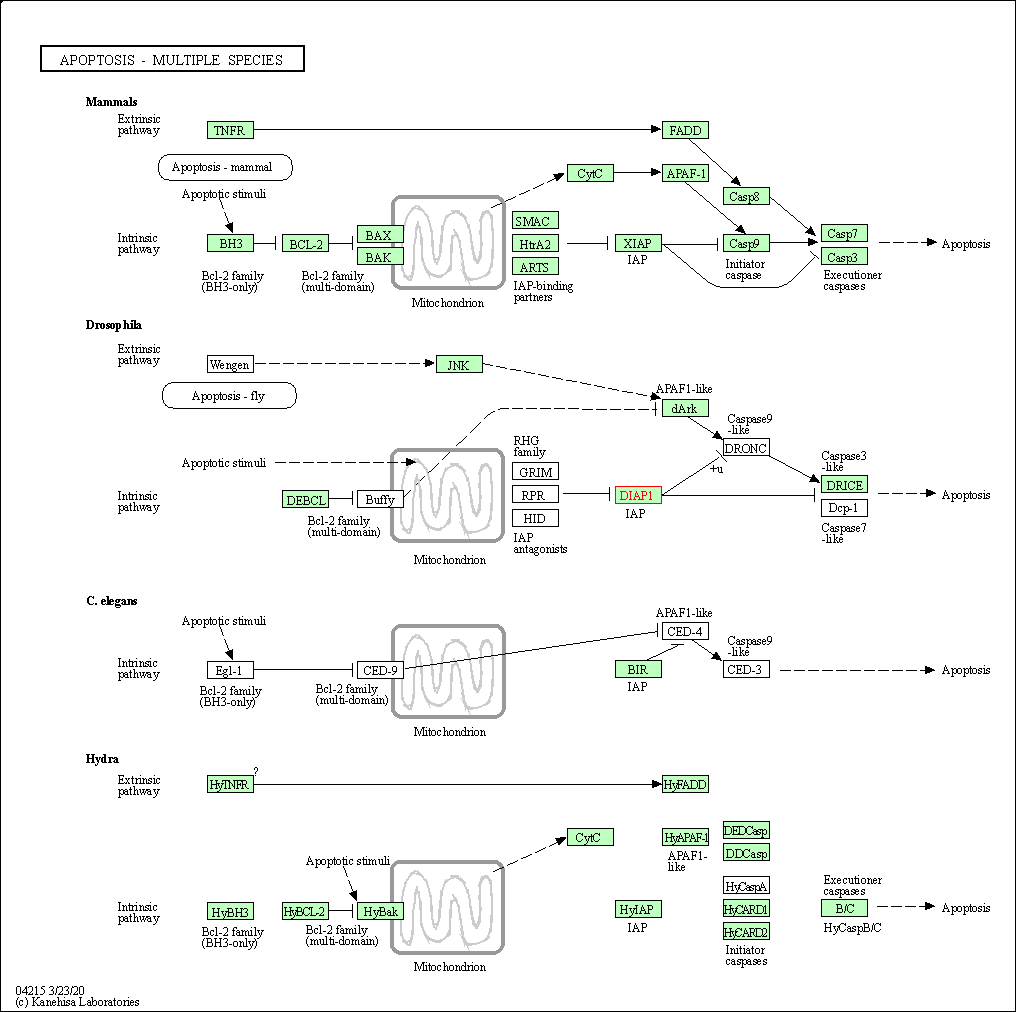
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ubiquitin mediated proteolysis | hsa04120 | Affiliated Target |

|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Apoptosis - multiple species | hsa04215 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 9.58E-06 |
|---|---|---|---|---|---|
| Closeness centrality | 2.12E-01 | Radiality | 1.37E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 2.78E+01 | Topological coefficient | 3.06E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Small molecule inhibitor of apoptosis proteins antagonists: a patent review.Expert Opin Ther Pat. 2015 Jul;25(7):755-74. | |||||
| REF 2 | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres. Bioorg Med Chem Lett. 2010 Apr 1;20(7):2229-33. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

