Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T43332
(Former ID: TTDR00947)
|
|||||
| Target Name |
Coagulation factor VII (F7)
|
|||||
| Synonyms |
Serum prothrombin conversion accelerator; SPCA; Proconvertin; Eptacog alfa
Click to Show/Hide
|
|||||
| Gene Name |
F7
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Coagulation defect [ICD-11: 3B10] | |||||
| Function |
Initiates the extrinsic pathway of blood coagulation. Serine protease that circulates in the blood in a zymogen form. Factor VII is converted to factor VIIa by factor Xa, factor XIIa, factor IXa, or thrombin by minor proteolysis. In the presence of tissue factor and calcium ions, factor VIIa then converts factor X to factor Xa by limited proteolysis. Factor VIIa will also convert factor IX to factor IXa in the presence of tissue factor and calcium.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.21.21
|
|||||
| Sequence |
MVSQALRLLCLLLGLQGCLAAGGVAKASGGETRDMPWKPGPHRVFVTQEEAHGVLHRRRR
ANAFLEELRPGSLERECKEEQCSFEEAREIFKDAERTKLFWISYSDGDQCASSPCQNGGS CKDQLQSYICFCLPAFEGRNCETHKDDQLICVNENGGCEQYCSDHTGTKRSCRCHEGYSL LADGVSCTPTVEYPCGKIPILEKRNASKPQGRIVGGKVCPKGECPWQVLLLVNGAQLCGG TLINTIWVVSAAHCFDKIKNWRNLIAVLGEHDLSEHDGDEQSRRVAQVIIPSTYVPGTTN HDIALLRLHQPVVLTDHVVPLCLPERTFSERTLAFVRFSLVSGWGQLLDRGATALELMVL NVPRLMTQDCLQQSRKVGDSPNITEYMFCAGYSDGSKDSCKGDSGGPHATHYRGTWYLTG IVSWGQGCATVGHFGVYTRVSQYIEWLQKLMRSEPRPGVLLRAPFP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Factor viia | Drug Info | Approved | Hemophilia | [2] | |
| Clinical Trial Drug(s) | [+] 7 Clinical Trial Drugs | + | ||||
| 1 | BAX-817 | Drug Info | Phase 3 | Hemophilia | [3] | |
| 2 | Vatreptacog alfa (activated) | Drug Info | Phase 3 | Bleeding disorder | [4] | |
| 3 | Activated recombinant FVII-albumin fusion protein | Drug Info | Phase 2/3 | Hemophilia | [5] | |
| 4 | Recombinant factor VIIa PEGylated liposomal | Drug Info | Phase 1/2 | Factor VII deficiency | [6] | |
| 5 | CB-813 | Drug Info | Phase 1 | Hemophilia | [7] | |
| 6 | Eptacog alfa | Drug Info | Phase 1 | Hemophilia | [8] | |
| 7 | F-7TG | Drug Info | Phase 1 | Hemophilia | [9] | |
| Discontinued Drug(s) | [+] 2 Discontinued Drugs | + | ||||
| 1 | BAY 86-6150 | Drug Info | Discontinued in Phase 2/3 | Hemophilia | [10] | |
| 2 | RNAPc2 | Drug Info | Discontinued in Phase 2 | Colorectal cancer | [11] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 13 Modulator drugs | + | ||||
| 1 | Factor viia | Drug Info | [1] | |||
| 2 | Vatreptacog alfa (activated) | Drug Info | [13] | |||
| 3 | Activated recombinant FVII-albumin fusion protein | Drug Info | [14] | |||
| 4 | Recombinant factor VIIa PEGylated liposomal | Drug Info | [1] | |||
| 5 | CB-813 | Drug Info | [15] | |||
| 6 | Eptacog alfa | Drug Info | [16] | |||
| 7 | F-7TG | Drug Info | [1] | |||
| 8 | BAY 86-6150 | Drug Info | [17] | |||
| 9 | RNAPc2 | Drug Info | [18] | |||
| 10 | Factor-VIIa-XTEN | Drug Info | [1] | |||
| 11 | Human recombinant factor VIIa | Drug Info | [1] | |||
| 12 | Long-acting Factor VII conjugate | Drug Info | [1] | |||
| 13 | MOD-5023 | Drug Info | [1] | |||
| Inhibitor | [+] 4 Inhibitor drugs | + | ||||
| 1 | BAX-817 | Drug Info | [12] | |||
| 2 | 4-(3,4-Diethoxy-benzylamino)-benzamidine | Drug Info | [19] | |||
| 3 | 4-(4-Benzyloxy-3-methoxy-benzylamino)-benzamidine | Drug Info | [19] | |||
| 4 | Fucose | Drug Info | [20] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Benzamidine | Ligand Info | |||||
| Structure Description | Cofactor-and substrate-assisted activation of factor VIIa | PDB:1KLI | ||||
| Method | X-ray diffraction | Resolution | 1.69 Å | Mutation | No | [21] |
| PDB Sequence |
IVGGKVCPKG
25 ECPWQVLLLV35 NGAQLCGGTL46 INTIWVVSAA56 HCFDKIKNWR62 NLIAVLGEHD 72 LSEHDGDEQS82 RRVAQVIIPS92 TYVPGTTNHD102 IALLRLHQPV112 VLTDHVVPLC 122 LPERTFSERT129C LAFVRFSLVS139 GWGQLLDRGA150 TALELMVLNV160 PRLMTQDCLQ 170 QSRKVGDSPN175 ITEYMFCAGY184 SDGSKDSCKG193 DSGGPHATHY203 RGTWYLTGIV 213 SWGQGCATVG223 HFGVYTRVSQ233 YIEWLQKLMR243 SEPRPGVLLR253 APFP |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Beta-D-Glucose | Ligand Info | |||||
| Structure Description | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla) Domains of Vitamin K-Dependent Clotting Factors At Physiological Ca2+ | PDB:3TH2 | ||||
| Method | X-ray diffraction | Resolution | 1.72 Å | Mutation | No | [22] |
| PDB Sequence |
ANAFLLRPGS
12 LRCKQCSFAR28 IFKDARTKLF40 WISYSDGDQC50 ASSPCQNGGS60 CKDQLQSYIC 70 FCLPAFEGRN80 CETHKDDQLI90 CVNENGGCEQ100 YCSDHTGTKR110 SCRCHEGYSL 120 LADGVSCTPT130 VEYPCGKIPI140 LE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
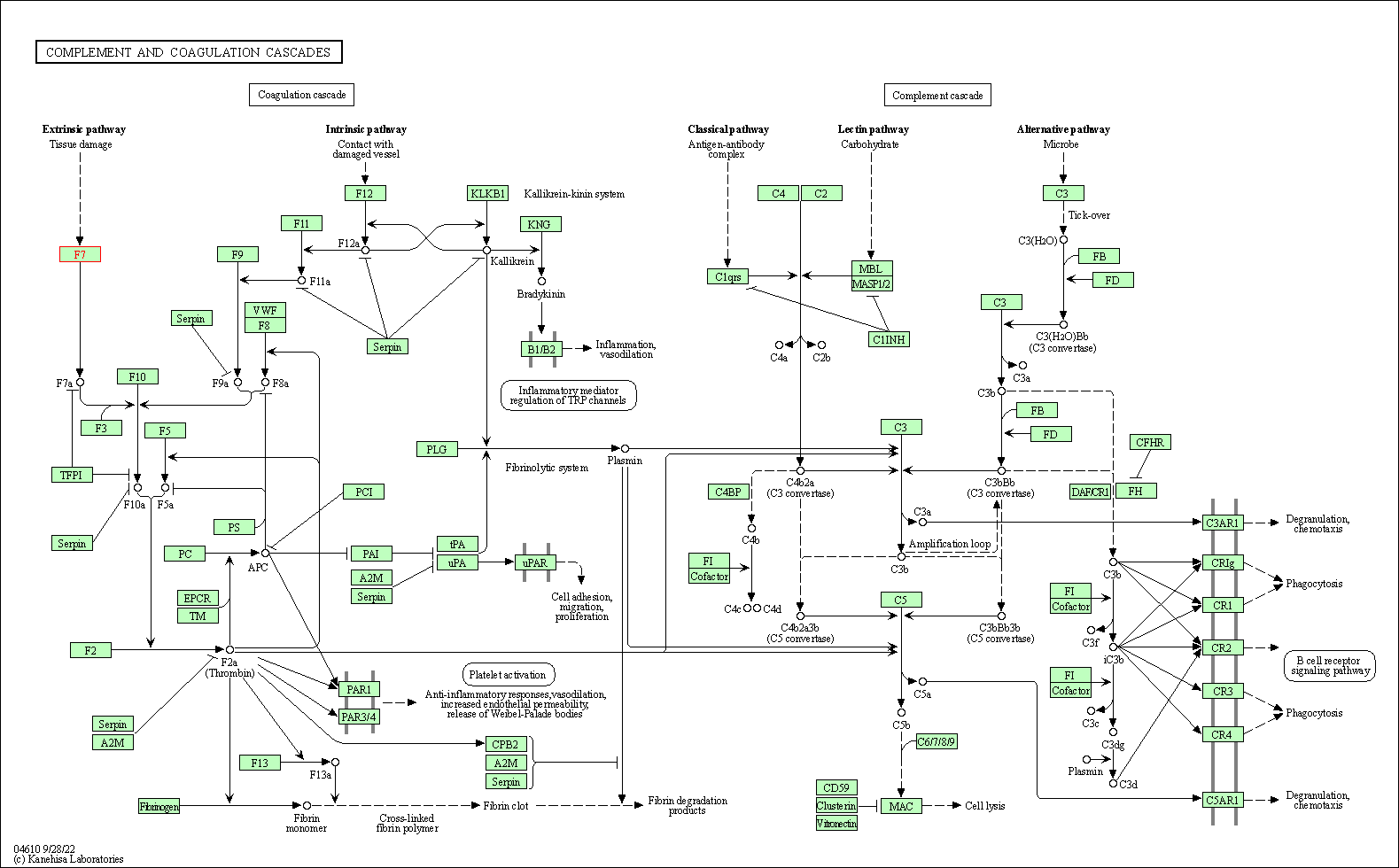
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Complement and coagulation cascades | hsa04610 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 6 | Degree centrality | 6.45E-04 | Betweenness centrality | 2.12E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.80E-01 | Radiality | 1.30E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 8.83E+00 | Topological coefficient | 2.80E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Complement and coagulation cascades | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Angiogenesis | |||||
| 2 | Blood coagulation | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Coagulation | |||||
| Reactome | [+] 5 Reactome Pathways | + | ||||
| 1 | BMAL1:CLOCK,NPAS2 activates circadian gene expression | |||||
| 2 | Extrinsic Pathway of Fibrin Clot Formation | |||||
| 3 | Gamma-carboxylation of protein precursors | |||||
| 4 | Transport of gamma-carboxylated protein precursors from the endoplasmic reticulum to the Golgi apparatus | |||||
| 5 | Removal of aminoterminal propeptides from gamma-carboxylated proteins | |||||
| WikiPathways | [+] 8 WikiPathways | + | ||||
| 1 | Complement and Coagulation Cascades | |||||
| 2 | PTM: gamma carboxylation, hypusine formation and arylsulfatase activation | |||||
| 3 | Blood Clotting Cascade | |||||
| 4 | Formation of Fibrin Clot (Clotting Cascade) | |||||
| 5 | Circadian Clock | |||||
| 6 | Folate Metabolism | |||||
| 7 | Vitamin B12 Metabolism | |||||
| 8 | Selenium Micronutrient Network | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2363). | |||||
| REF 2 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 3 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800032604) | |||||
| REF 4 | ClinicalTrials.gov (NCT01392547) Efficacy and Safety of NNC 0078-0000-0007 in Patients With Congenital Haemophilia and Inhibitors. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT02484638) Study of Recombinant Factor VIIa Fusion Protein (rVIIa-FP, CSL689) for On-demand Treatment of Bleeding Episodes in Patients With Hemophilia A or B With Inhibitors. | |||||
| REF 6 | Safety, pharmacokinetics and efficacy of factor VIIa formulated with PEGylated liposomes in haemophilia A patients with inhibitors to factor VIII--an open label, exploratory, cross-over, phase I/II study. Haemophilia. 2010 Nov;16(6):910-8. | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800027663) | |||||
| REF 8 | ClinicalTrials.gov (NCT02084810) Investigating the Bioequivalence of Eptacog Alfa A 6 mg and NovoSeven in Healthy Male Subjects. U.S. National Institutes of Health. | |||||
| REF 9 | GTC BIOTHERAPEUTICS REPORTS THIRD QUARTER 2010 FINANCIAL RESULTS. U.S. Securities and Exchange Commission. October 27, 2010. | |||||
| REF 10 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800021847) | |||||
| REF 11 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800006762) | |||||
| REF 12 | Clinical pipeline report, company report or official report of Baxter. | |||||
| REF 13 | Recombinant factor VIIa analog (vatreptacog alfa [activated]) for treatment of joint bleeds in hemophilia patients with inhibitors: a randomized controlled trial. J Thromb Haemost. 2012 Jan;10(1):81-9. | |||||
| REF 14 | ClinicalTrials.gov (NCT01542619) A Safety and Pharmacokinetics Study of a Recombinant Fusion Protein Linking Coagulation Factor VIIa With Albumin (rVIIa-FP) in Healthy Male Volunteers. U.S. National Institutes of Health. | |||||
| REF 15 | Clinical pipeline report, company report or official report of Catalyst Biosciences. | |||||
| REF 16 | Recombinant factor VIIa (eptacog alfa): a review of its use in congenital hemophilia with inhibitors, acquired hemophilia, and other congenital bleeding disorders. BioDrugs. 2008;22(2):121-36. | |||||
| REF 17 | Bayer HealthCare and Maxygen Announce Hematology Agreement. U.S. Securities and Exchange Commission. July 2, 2008. | |||||
| REF 18 | rNAPc2 inhibits colorectal cancer in mice through tissue factor. Clin Cancer Res. 2009 Jan 1;15(1):208-16. | |||||
| REF 19 | Design of selective phenylglycine amide tissue factor/factor VIIa inhibitors. Bioorg Med Chem Lett. 2005 Feb 1;15(3):817-22. | |||||
| REF 20 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 21 | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions. J Mol Biol. 2002 Sep 20;322(3):591-603. | |||||
| REF 22 | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla) | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

