Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T73908
(Former ID: TTDI02557)
|
|||||
| Target Name |
Wnt-7a protein (WNT7A)
|
|||||
| Synonyms |
Protein Wnt-7a
Click to Show/Hide
|
|||||
| Gene Name |
WNT7A
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Plays an important role in embryonic development, including dorsal versus ventral patterning during limb development, skeleton development and urogenital tract development. Required for central nervous system (CNS) angiogenesis and blood-brain barrier regulation. Required for normal, sexually dimorphic development of the Mullerian ducts, and for normal fertility in both sexes. Required for normal neural stem cell proliferation in the hippocampus dentate gyrus. Required for normal progress through the cell cycle in neural progenitor cells, for self-renewal of neural stem cells, and for normal neuronal differentiation and maturation. Promotes formation of synapses via its interaction with FZD5. Ligand for members of the frizzled family of seven transmembrane receptors that functions in the canonical Wnt/beta-catenin signaling pathway.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MNRKARRCLGHLFLSLGMVYLRIGGFSSVVALGASIICNKIPGLAPRQRAICQSRPDAII
VIGEGSQMGLDECQFQFRNGRWNCSALGERTVFGKELKVGSREAAFTYAIIAAGVAHAIT AACTQGNLSDCGCDKEKQGQYHRDEGWKWGGCSADIRYGIGFAKVFVDAREIKQNARTLM NLHNNEAGRKILEENMKLECKCHGVSGSCTTKTCWTTLPQFRELGYVLKDKYNEAVHVEP VRASRNKRPTFLKIKKPLSYRKPMDTDLVYIEKSPNYCEEDPVTGSVGTQGRACNKTAPQ ASGCDLMCCGRGYNTHQYARVWQCNCKFHWCCYVKCNTCSERTEMYTCK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Palmitoleic acid | Ligand Info | |||||
| Structure Description | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH O-PALMITOLEOYL SERINE - CRYSTAL FORM IX - 1.5A | PDB:4UZQ | ||||
| Method | X-ray diffraction | Resolution | 1.50 Å | Mutation | Yes | [2] |
| PDB Sequence |
S
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
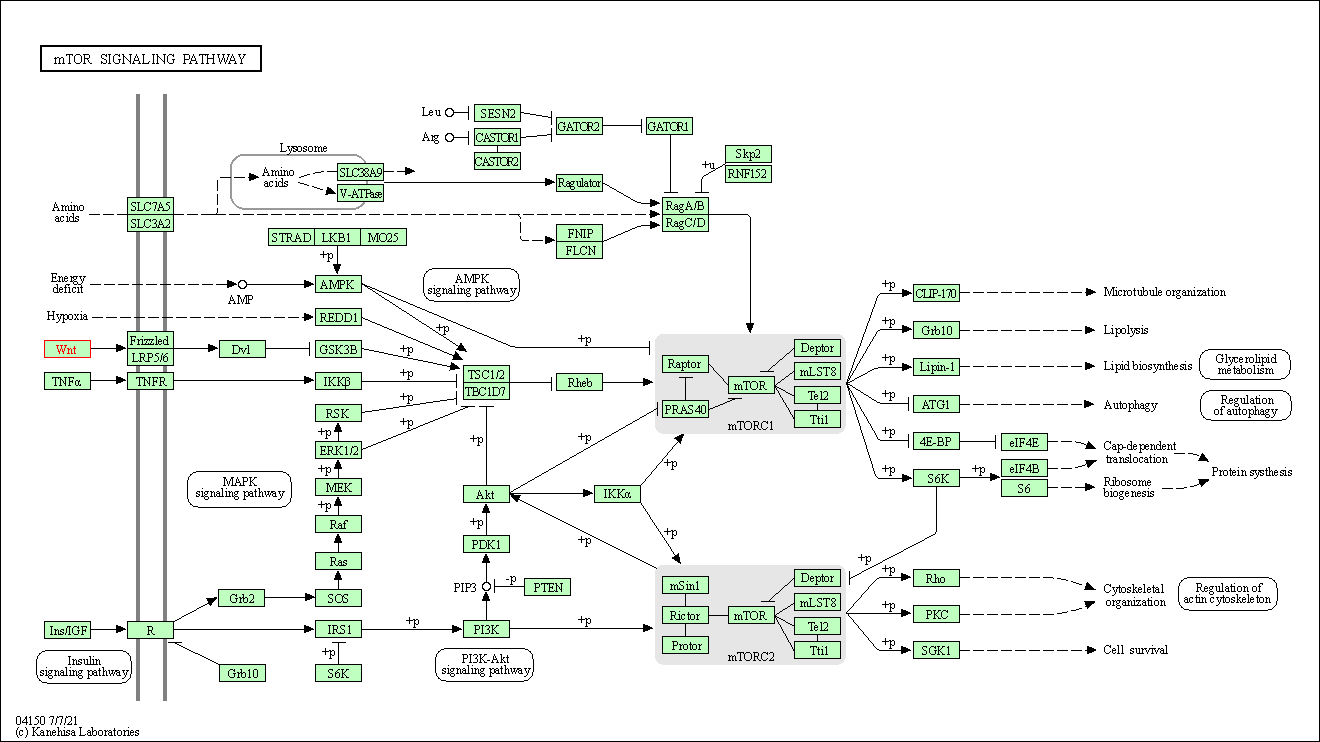
| mTOR signaling pathway | hsa04150 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
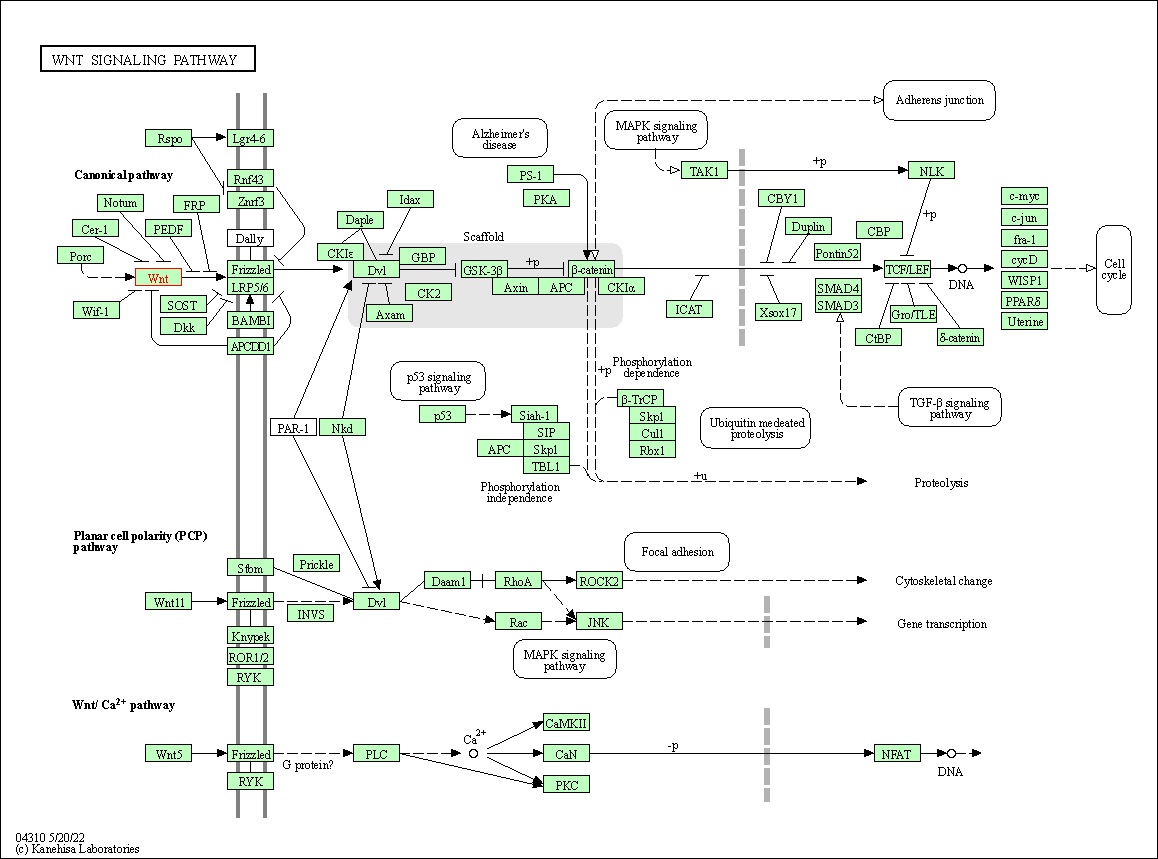
| Wnt signaling pathway | hsa04310 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
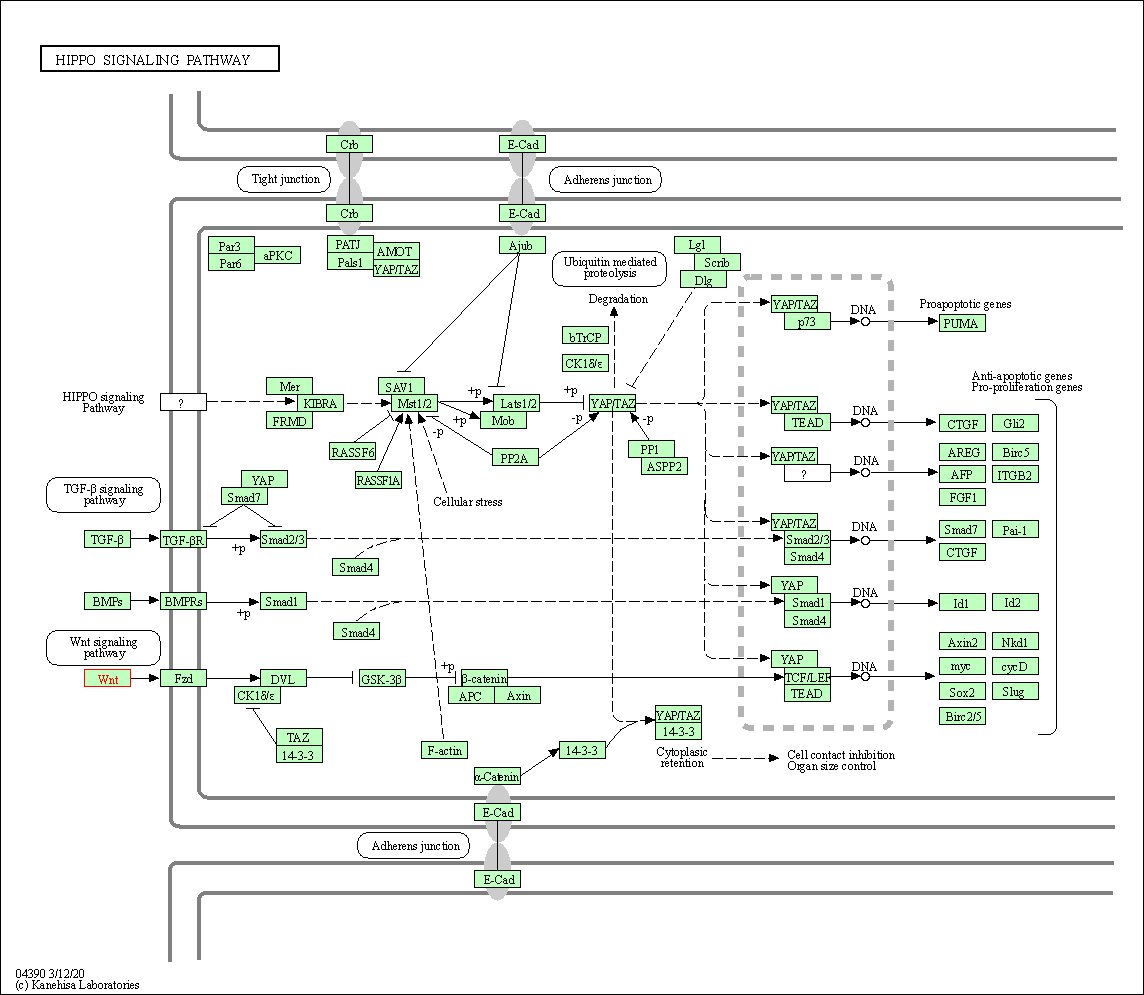
| Hippo signaling pathway | hsa04390 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
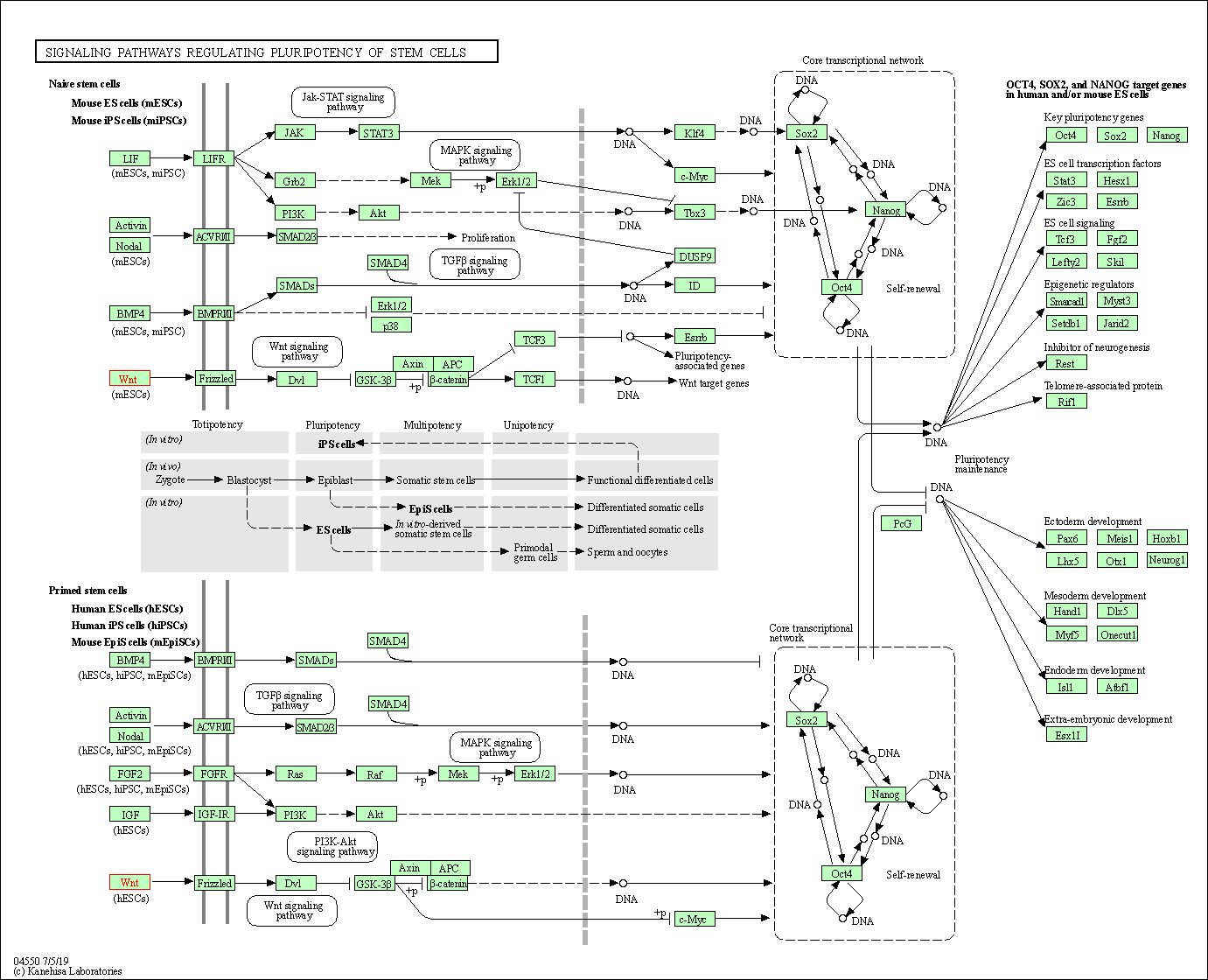
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Melanogenesis | hsa04916 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 4.41E-06 |
|---|---|---|---|---|---|
| Closeness centrality | 1.92E-01 | Radiality | 1.33E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 2.05E+01 | Topological coefficient | 3.75E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Wnt7a predicts poor prognosis, and contributes to growth and metastasis in tongue squamous cell carcinoma. Oncol Rep. 2019 Mar;41(3):1749-1758. | |||||
| REF 2 | Notum deacylates Wnt proteins to suppress signalling activity. Nature. 2015 Mar 12;519(7542):187-192. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

