Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T81623
(Former ID: TTDI03152)
|
|||||
| Target Name |
Death-associated protein kinase 3 (DAPK3)
|
|||||
| Synonyms |
Zipper-interacting protein kinase; ZIPK; ZIP-kinase; MYPT1 kinase; DAP-like kinase; DAP kinase 3
Click to Show/Hide
|
|||||
| Gene Name |
DAPK3
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Involved in the regulation of smooth muscle contraction. Regulates both type I (caspase-dependent) apoptotic and type II (caspase-independent) autophagic cell deaths signal, depending on the cellular setting. Involved in regulation of starvation-induced autophagy. Regulates myosin phosphorylation in both smooth muscle and non-muscle cells. In smooth muscle, regulates myosin either directly by phosphorylating MYL12B and MYL9 or through inhibition of smooth muscle myosin phosphatase (SMPP1M) via phosphorylation of PPP1R12A; the inhibition of SMPP1M functions to enhance muscle responsiveness to Ca(2+) and promote a contractile state. Phosphorylates MYL12B in non-muscle cells leading to reorganization of actin cytoskeleton. Isoform 2 can phosphorylate myosin, PPP1R12A and MYL12B. Overexpression leads to condensation of actin stress fibers into thick bundles. Involved in actin filament focal adhesion dynamics. The function in both reorganization of actin cytoskeleton and focal adhesion dissolution is modulated by RhoD. Positively regulates canonical Wnt/beta-catenin signaling through interaction with NLK and TCF7L2. Phosphorylates RPL13A on 'Ser-77' upon interferon-gamma activation which is causing RPL13A release from the ribosome, RPL13A association with the GAIT complex and its subsequent involvement in transcript-selective translation inhibition. Enhances transcription from AR-responsive promoters in a hormone- and kinase-dependent manner. Involved in regulation of cell cycle progression and cell proliferation. May be a tumor suppressor. Serine/threonine kinase which is involved in the regulation of apoptosis, autophagy, transcription, translation and actin cytoskeleton reorganization.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MSTFRQEDVEDHYEMGEELGSGQFAIVRKCRQKGTGKEYAAKFIKKRRLSSSRRGVSREE
IEREVNILREIRHPNIITLHDIFENKTDVVLILELVSGGELFDFLAEKESLTEDEATQFL KQILDGVHYLHSKRIAHFDLKPENIMLLDKNVPNPRIKLIDFGIAHKIEAGNEFKNIFGT PEFVAPEIVNYEPLGLEADMWSIGVITYILLSGASPFLGETKQETLTNISAVNYDFDEEY FSNTSELAKDFIRRLLVKDPKRRMTIAQSLEHSWIKAIRRRNVRGEDSGRKPERRRLKTT RLKEYTIKSHSSLPPNNSYADFERFSKVLEEAAAAEEGLRELQRSRRLCHEDVEALAAIY EEKEAWYREESDSLGQDLRRLRQELLKTEALKRQAQEEAKGALLGTSGLKRRFSRLENRY EALAKQVASEMRFVQDLVRALEQEKLQGVECGLR Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: CMP-6 | Ligand Info | |||||
| Structure Description | Crystal structure of human ZIP kinase in complex with a tetracyclic pyridone inhibitor (Pyridone 6) | PDB:2J90 | ||||
| Method | X-ray diffraction | Resolution | 2.00 Å | Mutation | No | [2] |
| PDB Sequence |
FQSMVEDHYE
14 MGEELGSGQF24 AIVRKCRQKG34 TGKEYAAKFI44 KKRRLSSRRG55 VSREEIEREV 65 NILREIRHPN75 IITLHDIFEN85 KTDVVLILEL95 VSGGELFDFL105 AEKESLTEDE 115 ATQFLKQILD125 GVHYLHSKRI135 AHFDLKPENI145 MLLDKNVPNP155 RIKLIDFGIA 165 HKIEGTPEFV184 APEIVNYEPL194 GLEADMWSIG204 VITYILLSGA214 SPFLGETKQE 224 TLTNISAVNY234 DFDEEYFSNT244 SELAKDFIRR254 LLVKDPKRRM264 IAQSLEHSWI 275 KAI
|
|||||
|
|
||||||
| Ligand Name: L-serine-O-phosphate | Ligand Info | |||||
| Structure Description | Crystal structure of human ZIP kinase in complex with a tetracyclic pyridone inhibitor (Pyridone 6) | PDB:2J90 | ||||
| Method | X-ray diffraction | Resolution | 2.00 Å | Mutation | No | [2] |
| PDB Sequence |
FQSMVEDHYE
14 MGEELGSGQF24 AIVRKCRQKG34 TGKEYAAKFI44 KKRRLSSRRG55 VSREEIEREV 65 NILREIRHPN75 IITLHDIFEN85 KTDVVLILEL95 VSGGELFDFL105 AEKESLTEDE 115 ATQFLKQILD125 GVHYLHSKRI135 AHFDLKPENI145 MLLDKNVPNP155 RIKLIDFGIA 165 HKIEGTPEFV184 APEIVNYEPL194 GLEADMWSIG204 VITYILLSGA214 SPFLGETKQE 224 TLTNISAVNY234 DFDEEYFSNT244 SELAKDFIRR254 LLVKDPKRRM264 IAQSLEHSWI 275 KAI
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
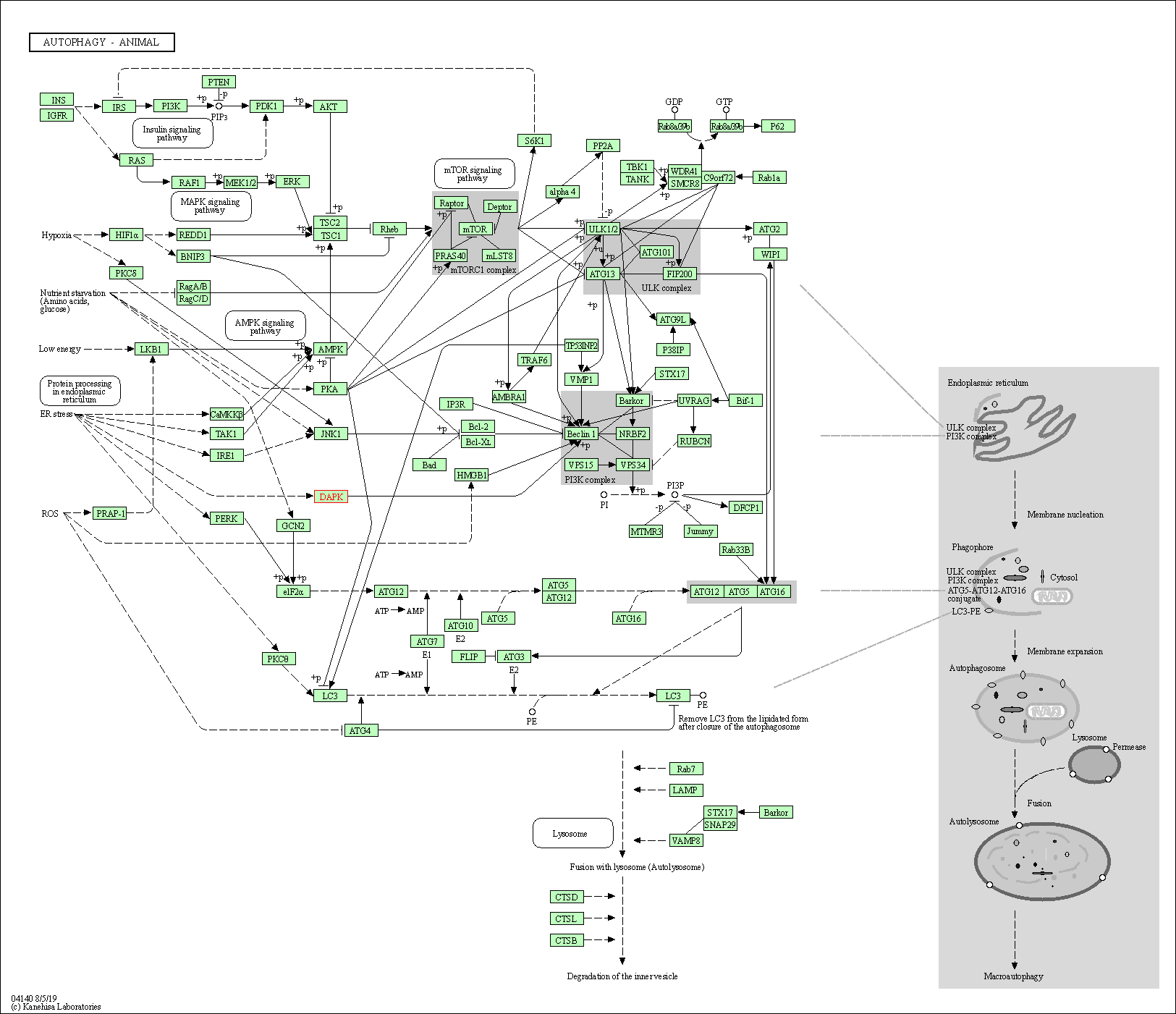
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Autophagy - animal | hsa04140 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Discovery, synthesis, and characterization of an orally bioavailable, brain penetrant inhibitor of mixed lineage kinase 3. J Med Chem. 2013 Oct 24;56(20):8032-48. | |||||
| REF 2 | Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J. 2008 Feb 20;27(4):704-14. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

