Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T95878
(Former ID: TTDI03522)
|
|||||
| Target Name |
Ribosomal protein S6 kinase alpha-6 (RSK6)
|
|||||
| Synonyms |
pp90RSK4; p90RSK6; p90-RSK 6; S6K-alpha-6; Ribosomal S6 kinase 4; RSK4; RSK-4; 90 kDa ribosomal protein S6 kinase 6
Click to Show/Hide
|
|||||
| Gene Name |
RPS6KA6
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Mature B-cell leukaemia [ICD-11: 2A82] | |||||
| Function |
Constitutively active serine/threonine-protein kinase that exhibits growth-factor-independent kinase activity and that may participate in p53/TP53-dependent cell growth arrest signaling and play an inhibitory role during embryogenesis.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MLPFAPQDEPWDREMEVFSGGGASSGEVNGLKMVDEPMEEGEADSCHDEGVVKEIPITHH
VKEGYEKADPAQFELLKVLGQGSFGKVFLVRKKTGPDAGQLYAMKVLKKASLKVRDRVRT KMERDILVEVNHPFIVKLHYAFQTEGKLYLILDFLRGGDVFTRLSKEVLFTEEDVKFYLA ELALALDHLHQLGIVYRDLKPENILLDEIGHIKLTDFGLSKESVDQEKKAYSFCGTVEYM APEVVNRRGHSQSADWWSYGVLMFEMLTGTLPFQGKDRNETMNMILKAKLGMPQFLSAEA QSLLRMLFKRNPANRLGSEGVEEIKRHLFFANIDWDKLYKREVQPPFKPASGKPDDTFCF DPEFTAKTPKDSPGLPASANAHQLFKGFSFVATSIAEEYKITPITSANVLPIVQINGNAA QFGEVYELKEDIGVGSYSVCKRCIHATTNMEFAVKIIDKSKRDPSEEIEILMRYGQHPNI ITLKDVFDDGRYVYLVTDLMKGGELLDRILKQKCFSEREASDILYVISKTVDYLHCQGVV HRDLKPSNILYMDESASADSIRICDFGFAKQLRGENGLLLTPCYTANFVAPEVLMQQGYD AACDIWSLGVLFYTMLAGYTPFANGPNDTPEEILLRIGNGKFSLSGGNWDNISDGAKDLL SHMLHMDPHQRYTAEQILKHSWITHRDQLPNDQPKRNDVSHVVKGAMVATYSALTHKTFQ PVLEPVAASSLAQRRSMKKRTSTGL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | BI-D1870 | Drug Info | Preclinical | Chronic lymphocytic leukaemia | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 4 Inhibitor drugs | + | ||||
| 1 | CID 138805970 | Drug Info | [2] | |||
| 2 | US10081632, Compound 25 | Drug Info | [2] | |||
| 3 | BI-D1870 | Drug Info | [1] | |||
| 4 | PMID22564207C25b | Drug Info | [3] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: AMP-PNP | Ligand Info | |||||
| Structure Description | RSK4 N-terminal Kinase Domain in complex with AMP-PNP | PDB:6G77 | ||||
| Method | X-ray diffraction | Resolution | 2.50 Å | Mutation | No | [4] |
| PDB Sequence |
VVKEIPITHH
60 VKEGYEKADP70 AQFELLKVLG80 QGSFGKVFLV90 RKKTGPDAGQ100 LYAMKVLKKA 110 SLKVRDDILV128 EVNHPFIVKL138 HYAFQTEGKL148 YLILDFLRGG158 DVFTRLSKEV 168 LFTEEDVKFY178 LAELALALDH188 LHQLGIVYRD198 LKPENILLDE208 IGHIKLTDFG 218 LSKESVDQEK228 KAYSFCGTVE238 YMAPEVVNRR248 GHSQSADWWS258 YGVLMFEMLT 268 GTLPFQGKDR278 NETMNMILKA288 KLGMPQFLSA298 EAQSLLRMLF308 KRNPANRLGS 318 EGVEEIKRHL328 FFANIDWDKL338 YKREVQPPFK348 P
|
|||||
|
|
VAL52
3.876
LEU79
3.718
GLY80
3.579
GLN81
3.749
GLY82
2.938
SER83
1.714
PHE84
2.640
GLY85
3.791
VAL87
3.277
ALA103
3.173
LYS105
2.664
LYS113
3.351
VAL114
4.147
ARG115
3.671
VAL136
4.210
LEU152
4.426
ASP153
2.642
PHE154
3.853
LEU155
3.154
ASP159
4.944
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: L-serine-O-phosphate | Ligand Info | |||||
| Structure Description | Phosphorylated RSK4 N-terminal Kinase Domain in complex with AMP-PNP | PDB:6G76 | ||||
| Method | X-ray diffraction | Resolution | 3.00 Å | Mutation | No | [4] |
| PDB Sequence |
VVKEIPITHH
60 VKEGYEKADP70 AQFELLKVLG80 QGSFGKVFLV90 RKKTGPDAGQ100 LYAMKVLKKA 110 SLKVRDDILV128 EVNHPFIVKL138 HYAFQTEGKL148 YLILDFLRGG158 DVFTRLSKEV 168 LFTEEDVKFY178 LAELALALDH188 LHQLGIVYRD198 LKPENILLDE208 IGHIKLTDFG 218 LSKESVDQEK228 KAYFCGTVEY239 MAPEVVNRRG249 HSQSADWWSY259 GVLMFEMLTG 269 TLPFQGKDRN279 ETMNMILKAK289 LGMPQFLSAE299 AQSLLRMLFK309 RNPANRLGSE 319 GVEEIKRHLF329 FANIDWDKLY339 KREVQPPFKP349
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
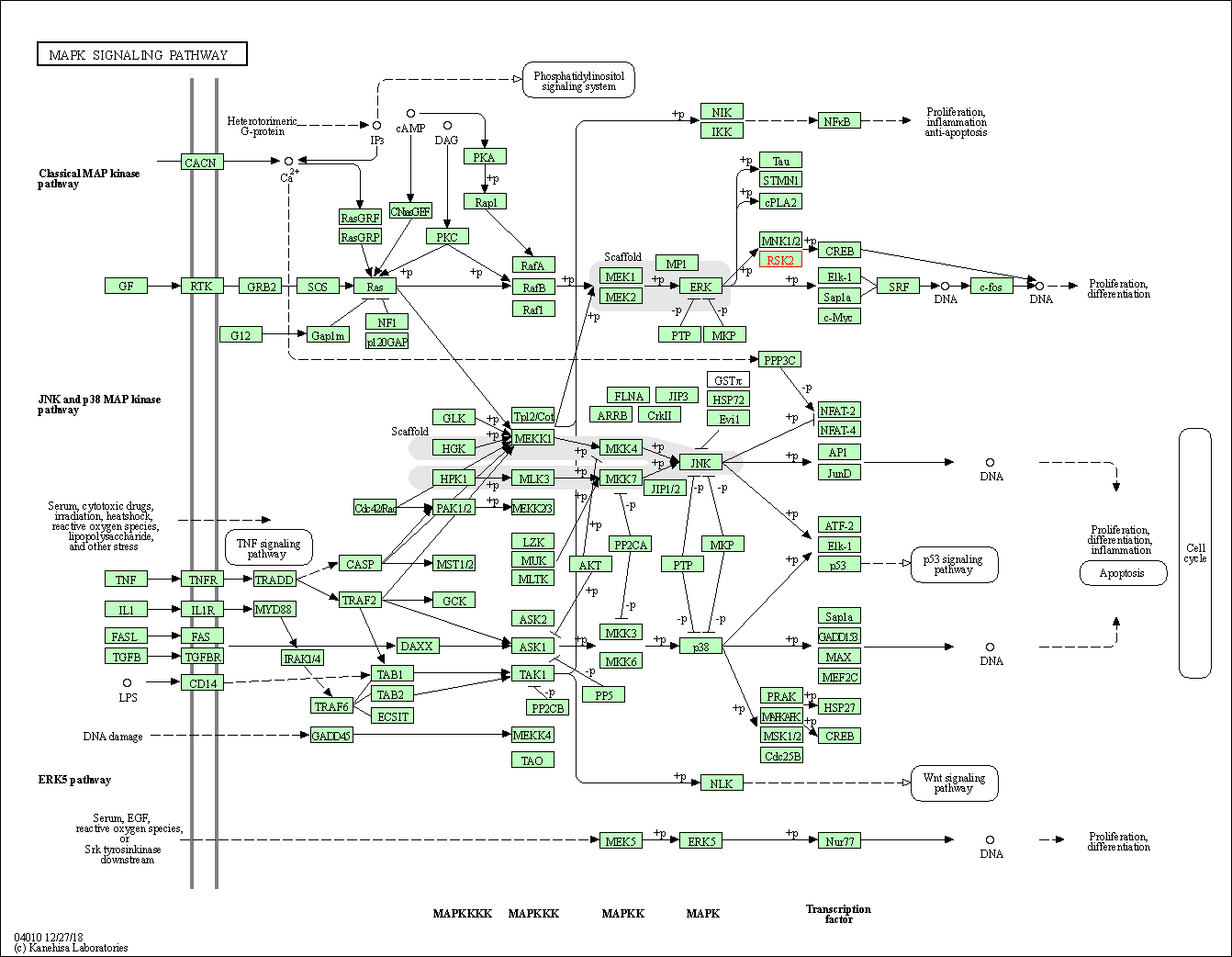
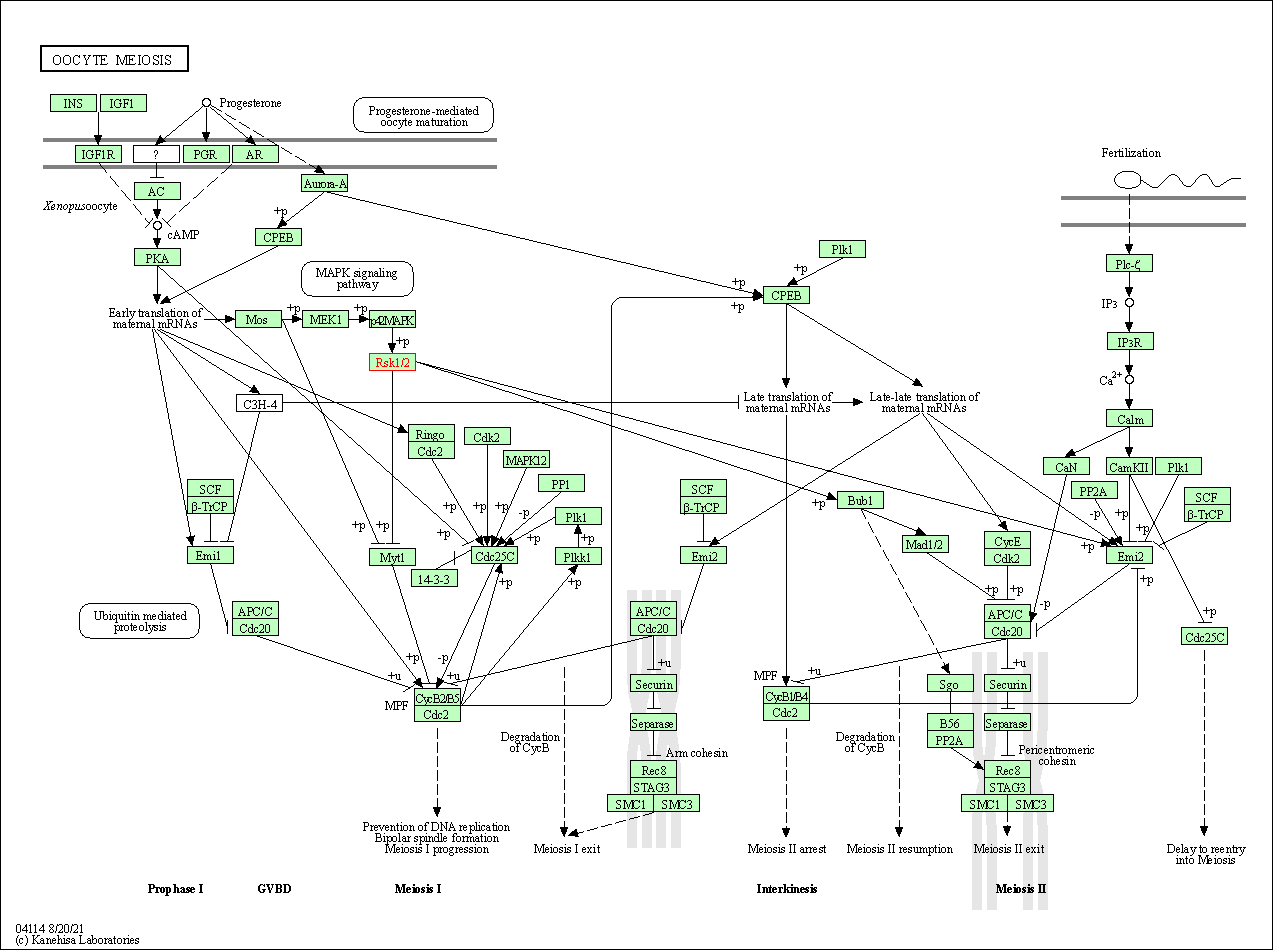
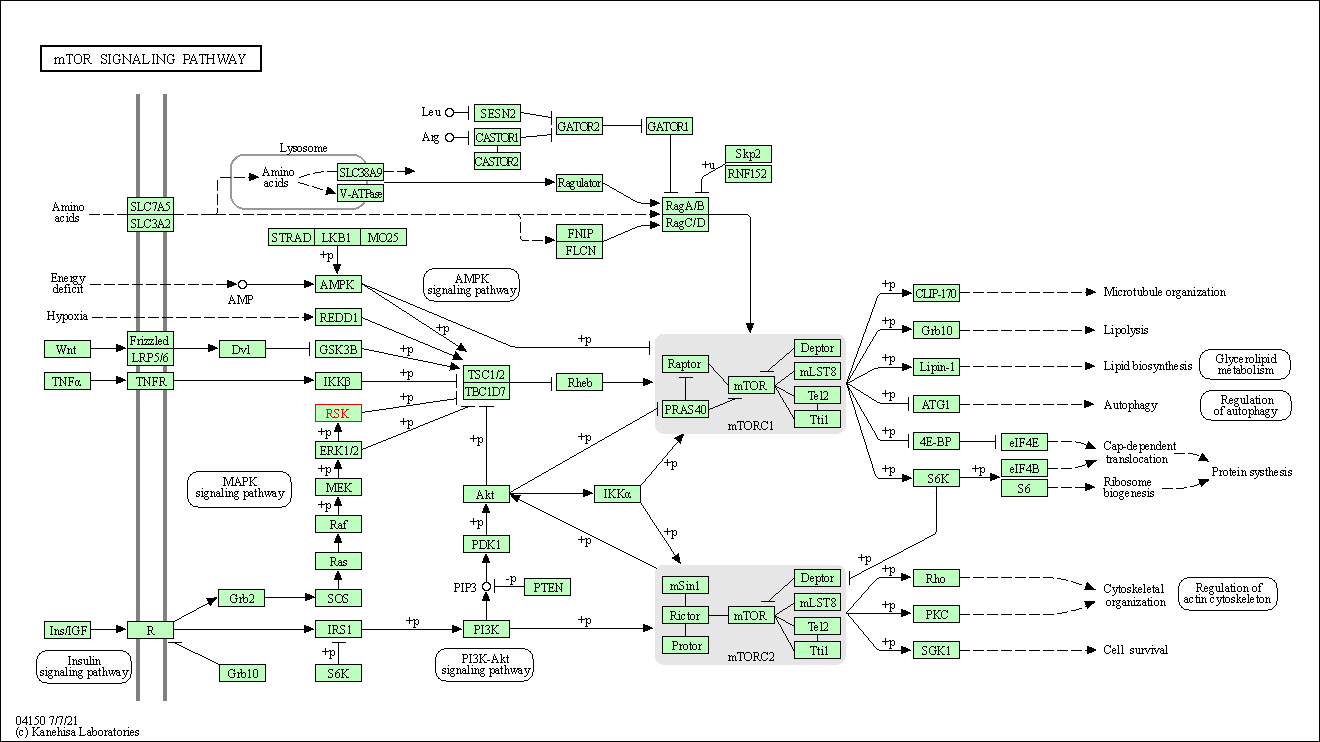
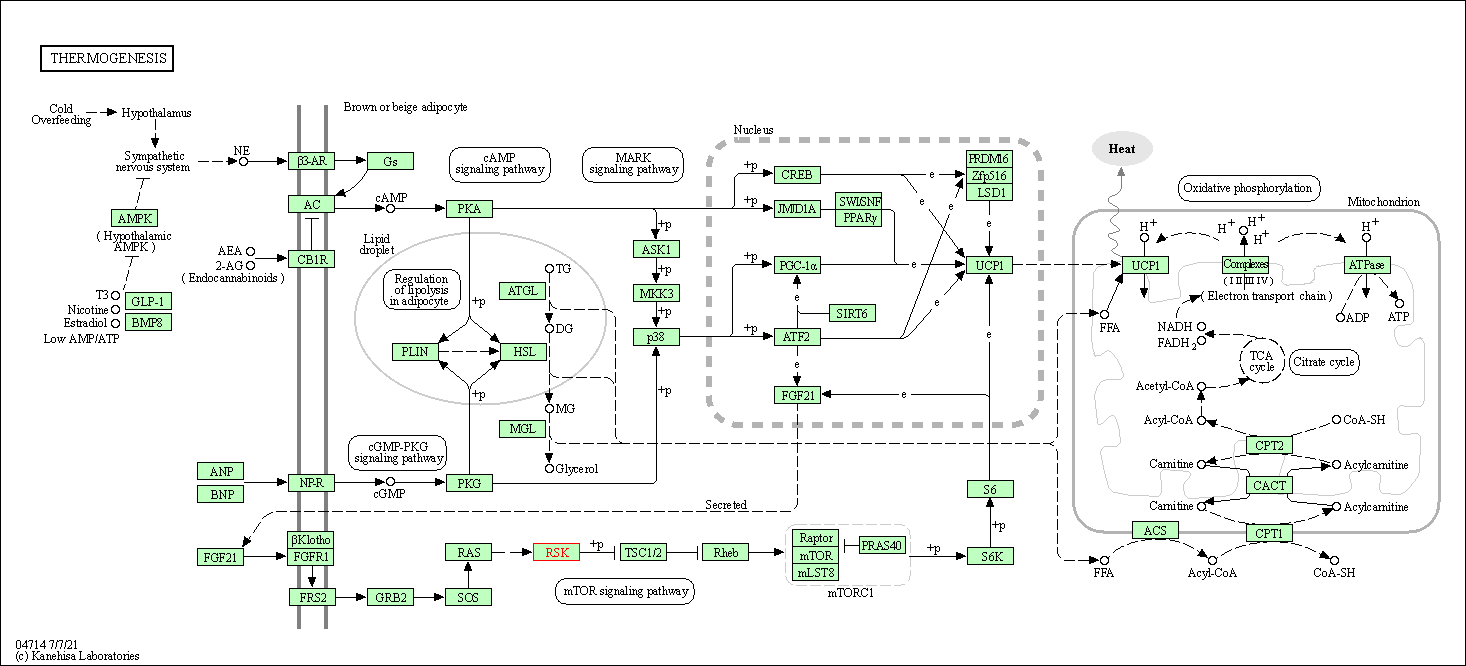
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| mTOR signaling pathway | hsa04150 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Thermogenesis | hsa04714 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Long-term potentiation | hsa04720 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J. 2007 Jan 1;401(1):29-38. | |||||
| REF 2 | Substituted tetrahydropyrido[3,2:4,5]pyrrolo[1,2-a]pyrazine-2-carboxamides as RSK inhibitors. US9771366. | |||||
| REF 3 | Discovery of an orally efficacious inhibitor of anaplastic lymphoma kinase. J Med Chem. 2012 May 24;55(10):4580-93. | |||||
| REF 4 | Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer. Sci Transl Med. 2021 Jul 14;13(602):eaba4627. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

