Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T97537
(Former ID: TTDI03348)
|
|||||
| Target Name |
Leucyl-cysteinyl aminopeptidase (LNPEP)
|
|||||
| Synonyms |
Placental leucine aminopeptidase; P-LAP; Oxytocinase; OTase; Leucyl-cystinyl aminopeptidase; Insulin-responsive aminopeptidase; Insulin-regulated membrane aminopeptidase; IRAP; Cystinyl aminopeptidase
Click to Show/Hide
|
|||||
| Gene Name |
LNPEP
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Function |
Degrades peptide hormones such as oxytocin, vasopressin and angiotensin III, and plays a role in maintaining homeostasis during pregnancy. May be involved in the inactivation of neuronal peptides in the brain. Cleaves Met-enkephalin and dynorphin. Binds angiotensin IV and may be the angiotensin IV receptor in the brain. Release of an N-terminal amino acid, cleaves before cysteine, leucine as well as other amino acids.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.11.3
|
|||||
| Sequence |
MEPFTNDRLQLPRNMIENSMFEEEPDVVDLAKEPCLHPLEPDEVEYEPRGSRLLVRGLGE
HEMEEDEEDYESSAKLLGMSFMNRSSGLRNSATGYRQSPDGACSVPSARTMVVCAFVIVV AVSVIMVIYLLPRCTFTKEGCHKKNQSIGLIQPFATNGKLFPWAQIRLPTAVVPLRYELS LHPNLTSMTFRGSVTISVQALQVTWNIILHSTGHNISRVTFMSAVSSQEKQAEILEYAYH GQIAIVAPEALLAGHNYTLKIEYSANISSSYYGFYGFSYTDESNEKKYFAATQFEPLAAR SAFPCFDEPAFKATFIIKIIRDEQYTALSNMPKKSSVVLDDGLVQDEFSESVKMSTYLVA FIVGEMKNLSQDVNGTLVSIYAVPEKIGQVHYALETTVKLLEFFQNYFEIQYPLKKLDLV AIPDFEAGAMENWGLLTFREETLLYDSNTSSMADRKLVTKIIAHELAHQWFGNLVTMKWW NDLWLNEGFATFMEYFSLEKIFKELSSYEDFLDARFKTMKKDSLNSSHPISSSVQSSEQI EEMFDSLSYFKGSSLLLMLKTYLSEDVFQHAVVLYLHNHSYASIQSDDLWDSFNEVTNQT LDVKRMMKTWTLQKGFPLVTVQKKGKELFIQQERFFLNMKPEIQPSDTSYLWHIPLSYVT EGRNYSKYQSVSLLDKKSGVINLTEEVLWVKVNINMNGYYIVHYADDDWEALIHQLKINP YVLSDKDRANLINNIFELAGLGKVPLKRAFDLINYLGNENHTAPITEALFQTDLIYNLLE KLGYMDLASRLVTRVFKLLQNQIQQQTWTDEGTPSMRELRSALLEFACTHNLGNCSTTAM KLFDDWMASNGTQSLPTDVMTTVFKVGAKTDKGWSFLLGKYISIGSEAEKNKILEALASS EDVRKLYWLMKSSLNGDNFRTQKLSFIIRTVGRHFPGHLLAWDFVKENWNKLVQKFPLGS YTIQNIVAGSTYLFSTKTHLSEVQAFFENQSEATFRLRCVQEALEVIQLNIQWMEKNLKS LTWWL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: L-lysine | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Insulin Regulated Aminopeptidase with Lysine in Active Site | PDB:4PJ6 | ||||
| Method | X-ray diffraction | Resolution | 2.96 Å | Mutation | No | [5] |
| PDB Sequence |
KLFPWAQIRL
168 PTAVVPLRYE178 LSLHPNLTSM188 TFRGSVTISV198 QALQVTWNII208 LHSTGHNISR 218 VTFMSVSSQE229 KQAEILEYAY239 HGQIAIVAPE249 ALLAGHNYTL259 KIEYSANISS 269 SYYGFYGFSY279 TDESNEKKYF289 AATQFEPLAA299 RSAFPCFDEP309 AFKATFIIKI 319 IRDEQYTALS329 NMPKKSSVVL339 DDGLVQDEFS349 ESVKMSTYLV359 AFIVGEMKNL 369 SQDVNGTLVS379 IYAVPEKIGQ389 VHYALETTVK399 LLEFFQNYFE409 IQYPLKKLDL 419 VAIPDFEAGA429 MENWGLLTFR439 EETLLYDSNT449 SSMADRKLVT459 KIIAHELAHQ 469 WFGNLVTMKW479 WNDLWLNEGF489 ATFMEYFSLE499 KIFKELSSYE509 DFLDARFKTM 519 KKDSLNSSHP529 ISSSVQSSEQ539 IEEMFDSLSY549 FKGSSLLLML559 KTYLSEDVFQ 569 HAVVLYLHNH579 SYASIQSDDL589 WDSFNEVTNQ599 TLDVKRMMKT609 WTLQKGFPLV 619 TVQKKGKELF629 IQQERFFLNM639 SDTSYLWHIP655 LSYVTEGRNY665 SKYQSVSLLD 675 KKSGVINLTE685 EVLWVKVNIN695 MNGYYIVHYA705 DDDWEALIHQ715 LKINPYVLSD 725 KDRANLINNI735 FELAGLGKVP745 LKRAFDLINY755 LGNENHTAPI765 TEALFQTDLI 775 YNLLEKLGYM785 DLASRLVTRV795 FKLLQNQIQQ805 QTWTDEGTPS815 MRELRSALLE 825 FACTHNLGNC835 STTAMKLFDD845 WMASNGTQSL855 PTDVMTTVFK865 VGAKTDKGWS 875 FLLGKYISIG885 SEAEKNKILE895 ALASSEDVRK905 LYWLMKSSLN915 GDNFRTQKLS 925 FIIRTVGRHF935 PGHLLAWDFV945 KENWNKLVQK955 FPLGSYTIQN965 IVAGSTYLFS 975 TKTHLSEVQA985 FFENQSEATF995 RLRCVQEALE1005 VIQLNIQWME1015 KNLKSLTWWL 1025
|
|||||
|
|
||||||
| Ligand Name: [(2~{s})-2-[[(2~{s})-1-Azanyl-1-Oxidanylidene-3-Phenyl-Propan-2-Yl]carbamoyl]-4,4-Diphenyl-Butyl]-[(1~{r})-1-Azanyl-3-Phenyl-Propyl]phosphinic Acid | Ligand Info | |||||
| Structure Description | Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. | PDB:5MJ6 | ||||
| Method | X-ray diffraction | Resolution | 2.53 Å | Mutation | No | [6] |
| PDB Sequence |
NGKLFPWAQI
166 RLPTAVVPLR176 YELSLHPNLT186 SMTFRGSVTI196 SVQALQVTWN206 IILHSTGHNI 216 SRVTFMSAVS226 SQEKQAEILE236 YAYHGQIAIV246 APEALLAGHN256 YTLKIEYSAN 266 ISSSYYGFYG276 FSYTDESNEK286 KYFAATQFEP296 LAARSAFPCF306 DEPAFKATFI 316 IKIIRDEQYT326 ALSNMPKKSS336 VVLDDGLVQD346 EFSESVKMST356 YLVAFIVGEM 366 KNLSQDVNGT376 LVSIYAVPEK386 IGQVHYALET396 TVKLLEFFQN406 YFEIQYPLKK 416 LDLVAIPDFE426 AGAMENWGLL436 TFREETLLYD446 SNTSSMADRK456 LVTKIIAHEL 466 AHQWFGNLVT476 MKWWNDLWLN486 EGFATFMEYF496 SLEKIFKELS506 SYEDFLDARF 516 KTMKKDSLNS526 SHPISSSVQS536 SEQIEEMFDS546 LSYFKGSSLL556 LMLKTYLSED 566 VFQHAVVLYL576 HNHSYASIQS586 DDLWDSFNEV596 TNQTLDVKRM606 MKTWTLQKGF 616 PLVTVQKKGK626 ELFIQQERFF636 LNMSYLWHIP655 LSYVTEGRNY665 SKYQSVSLLD 675 KKSGVINLTE685 EVLWVKVNIN695 MNGYYIVHYA705 DDDWEALIHQ715 LKINPYVLSD 725 KDRANLINNI735 FELAGLGKVP745 LKRAFDLINY755 LGNENHTAPI765 TEALFQTDLI 775 YNLLEKLGYM785 DLASRLVTRV795 FKLLQNQIQQ805 QTWTDEGTPS815 MRELRSALLE 825 FACTHNLGNC835 STTAMKLFDD845 WMASNGTQSL855 PTDVMTTVFK865 VGAKTDKGWS 875 FLLGKYISIG885 SEAEKNKILE895 ALASSEDVRK905 LYWLMKSSLN915 GDNFRTQKLS 925 FIIRTVGRHF935 PGHLLAWDFV945 KENWNKLVQK955 FPLGSYTIQN965 IVAGSTYLFS 975 TKTHLSEVQA985 FFENQSEATF995 RLRCVQEALE1005 VIQLNIQWME1015 KNLKSLTWWL 1025 RTETSQVAPA1035
|
|||||
|
|
GLN293
4.303
GLU295
2.728
PRO296
4.507
GLU426
4.857
ALA427
3.665
GLY428
2.990
ALA429
3.231
MET430
3.883
GLU431
2.741
LEU457
3.309
LYS460
4.100
ILE461
3.515
HIS464
3.254
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
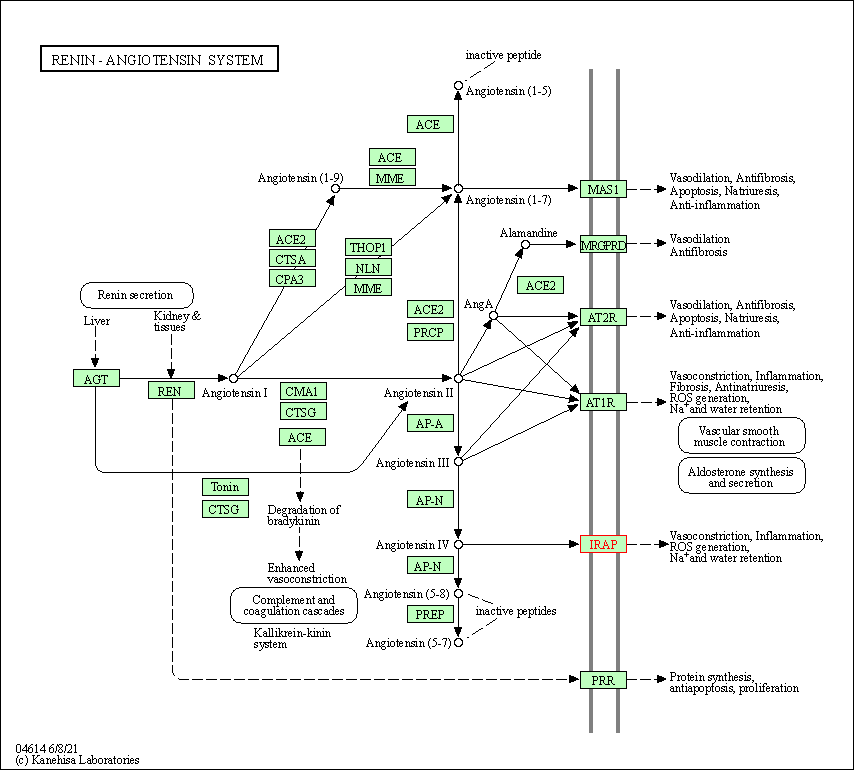
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Renin-angiotensin system | hsa04614 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 6 | Degree centrality | 6.45E-04 | Betweenness centrality | 4.46E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.97E-01 | Radiality | 1.34E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 8.33E+00 | Topological coefficient | 2.16E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Potent macrocyclic inhibitors of insulin-regulated aminopeptidase (IRAP) by olefin ring-closing metathesis. J Med Chem. 2011 Jun 9;54(11):3779-92. | |||||
| REF 2 | Bicyclic pyridine compound. US10023583. | |||||
| REF 3 | Pyridine derivative. US10059720. | |||||
| REF 4 | Novel selective inhibitors of aminopeptidases that generate antigenic peptides. Bioorg Med Chem Lett. 2013 Sep 1;23(17):4832-6. | |||||
| REF 5 | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides. Protein Sci. 2015 Feb;24(2):190-9. | |||||
| REF 6 | Ligand-Induced Conformational Change of Insulin-Regulated Aminopeptidase: Insights on Catalytic Mechanism and Active Site Plasticity. J Med Chem. 2017 Apr 13;60(7):2963-2972. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

