| Drug General Information |
| Drug ID |
D08CDI
|
| Former ID |
DAP000853
|
| Drug Name |
Lapatinib
|
| Synonyms |
FMM; Tycerb; Lapatinib Ditosylate; Lapatinib [INN]; Lapatinib tosilate hydrate; GSK 572016; GSK572016; GW 572016; GW 572016X; GW572016; Lapatinib (INN); Tykerb (TN); Lapatinib, Tykerb, GW572016; N-[3-chloro-4-[(3-fluorophenyl)methoxy]phenyl]-6-[5-[(2-methylsulfonylethylamino)methyl]furan-2-yl]quinazolin-4-amine; N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE; N-(3-Chloro-4-((3-fluorophenyl)methoxy)phenyl)-6-(5-((2-methylsulfonylethylamino)methyl)-2-furyl)quinazolin-4-amine; N-(3-Chloro-4-{[(3-fluorophenyl)methyl]oxy}phenyl)-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furanyl]-4-quinazolinamine; 4-[[3-Chloro-4-(3-fluorobenzyloxy)phenyl]amino]-6-[5-[[(2-methanesulfonylethyl)amino]methyl]furan-2-yl]quinazoline; Lapatinib (ERBB2 inhibitor)
|
| Drug Type |
Small molecular drug
|
| Therapeutic Class |
Anticancer Agents
|
| Company |
GlaxoSmithKline
|
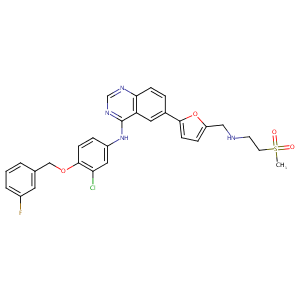
| Structure |
|
Download
2D MOL
3D MOL
|
| Formula |
C29H26ClFN4O4S
|
| InChI |
InChI=1S/C29H26ClFN4O4S/c1-40(36,37)12-11-32-16-23-7-10-27(39-23)20-5-8-26-24(14-20)29(34-18-33-26)35-22-6-9-28(25(30)15-22)38-17-19-3-2-4-21(31)13-19/h2-10,13-15,18,32H,11-12,16-17H2,1H3,(H,33,34,35)
|
| InChIKey |
BCFGMOOMADDAQU-UHFFFAOYSA-N
|
| CAS Number |
CAS 388082-78-8
|
| PubChem Compound ID |
|
| PubChem Substance ID |
585695, 7887520, 8035064, 9368726, 14911387, 21317859, 30413551, 46393564, 46506302, 46507141, 49742619, 50070568, 50071307, 50100107, 50112760, 50644701, 53788364, 57399558, 85171071, 85202079, 91147938, 92308826, 92719029, 93581028, 96024798, 103177479, 103854383, 103905567, 103905568, 109692966, 113442073, 117695459, 124360113, 124893335, 124893336, 125345521, 126592984, 126621155, 126649062, 126666978, 126667073, 126731332, 127325943, 127325944, 127325945, 127494626, 134338132, 135128225, 135685383, 135685387
|
| SuperDrug ATC ID |
L01XE07
|
| Drug Resistance Mutation (DRM) |
| Target and Pathway |
| References |