| Drug General Information |
| Drug ID |
D0D1GF
|
| Former ID |
DCL000265
|
| Drug Name |
XL880
|
| Synonyms |
GSK 089; GSK 1363089; GSK1363089; XL 880; GSK1363089, GSK089, foretinib, EXEL-2880, XL880; 88Z; MET inhibitors
|
| Drug Type |
Small molecular drug
|
| Indication |
Gastric cancer; Renal cell carcinoma; Squamous cell cancer of head and neck [ICD9: 140-229, 151, 189; ICD10:C16, C64]
|
Phase 2 |
[1],
[2]
|
|---|
|
Solid tumours [ICD9: 140-199, 210-229; ICD10:C00-D48]
|
Phase 1 |
[1],
[2]
|
| Therapeutic Class |
Anticancer Agents
|
| Company |
Exelixis; GlaxoSmithKline
|
| Structure |
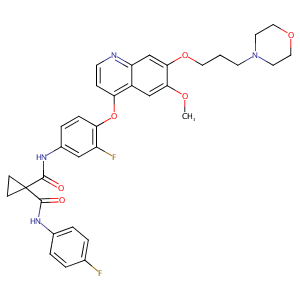
|
Download
2D MOL
3D MOL
|
| Formula |
C34H34F2N4O6
|
| InChI |
InChI=1S/C34H34F2N4O6/c1-43-30-20-25-27(21-31(30)45-16-2-13-40-14-17-44-18-15-40)37-12-9-28(25)46-29-8-7-24(19-26(29)36)39-33(42)34(10-11-34)32(41)38-23-5-3-22(35)4-6-23/h3-9,12,19-21H,2,10-11,13-18H2,1H3,(H,38,41)(H,39,42)
|
| InChIKey |
CXQHYVUVSFXTMY-UHFFFAOYSA-N
|
| PubChem Compound ID |
|
| PubChem Substance ID |
81082087, 86465340, 93375504, 99436987, 104246801, 123055389, 124490358, 124757009, 125163814, 125329934, 126731542, 131480687, 134338922, 134964398, 135257201, 135727454, 136367309, 136367948, 136920382, 137245315, 137275745, 137275948, 137760554, 143499560, 144115968, 152043738, 152234432, 152258182, 152344218, 160647018, 162011882, 162037444, 162196274, 163348564, 164041883, 170501826, 172913907, 174530915, 177748909, 178102306, 185997073, 185998158, 186000980, 188899550, 198948852, 198993146, 223387910, 223704673, 224083490, 226703719
|
| Target and Pathway |
| Target(s) |
Vascular endothelial growth factor receptor 2 |
Target Info |
Modulator |
[3]
|
|---|
| Hepatocyte growth factor receptor |
Target Info |
Modulator |
[3]
|
|
KEGG Pathway
|
Ras signaling pathway
|
|
Rap1 signaling pathway
|
|
Cytokine-cytokine receptor interaction
|
|
Endocytosis
|
|
PI3K-Akt signaling pathway
|
|
VEGF signaling pathway
|
|
Focal adhesion
|
|
Proteoglycans in cancerhsa04014:Ras signaling pathway
|
|
Axon guidance
|
|
Adherens junction
|
|
Bacterial invasion of epithelial cells
|
|
Epithelial cell signaling in Helicobacter pylori infection
|
|
Malaria
|
|
Pathways in cancer
|
|
Transcriptional misregulation in cancer
|
|
Proteoglycans in cancer
|
|
MicroRNAs in cancer
|
|
Renal cell carcinoma
|
|
Melanoma
|
|
Central carbon metabolism in cancer
|
|
NetPath Pathway
|
IL2 Signaling Pathway
|
|
PANTHER Pathway
|
Angiogenesis
|
|
VEGF signaling pathway
|
|
Pathway Interaction Database
|
HIF-2-alpha transcription factor network
|
|
Beta3 integrin cell surface interactions
|
|
Signaling events mediated by TCPTP
|
|
SHP2 signaling
|
|
S1P1 pathway
|
|
VEGF and VEGFR signaling network
|
|
Integrins in angiogenesis
|
|
Signaling events mediated by VEGFR1 and VEGFR2
|
|
Notch-mediated HES/HEY networkmet_pathway:Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met)
|
|
Arf6 signaling events
|
|
Posttranslational regulation of adherens junction stability and dissassembly
|
|
Direct p53 effectors
|
|
Syndecan-1-mediated signaling events
|
|
Stabilization and expansion of the E-cadherin adherens junction
|
|
a6b1 and a6b4 Integrin signaling
|
|
FGF signaling pathway
|
|
Regulation of retinoblastoma protein
|
|
Reactome
|
Neurophilin interactions with VEGF and VEGFR
|
|
VEGF binds to VEGFR leading to receptor dimerization
|
|
Integrin cell surface interactions
|
|
EPHA-mediated growth cone collapse
|
|
VEGFA-VEGFR2 Pathway
|
|
VEGFR2 mediated cell proliferationR-HSA-416550:Sema4D mediated inhibition of cell attachment and migration
|
|
WikiPathways
|
Focal Adhesion
|
|
Nifedipine Activity
|
|
Cardiac Progenitor Differentiation
|
|
Signaling by VEGF
|
|
AngiogenesisWP366:TGF beta Signaling Pathway
|
|
Signaling of Hepatocyte Growth Factor Receptor
|
|
Extracellular vesicle-mediated signaling in recipient cells
|
|
Signaling Pathways in Glioblastoma
|
|
miR-targeted genes in squamous cell - TarBase
|
|
miR-targeted genes in muscle cell - TarBase
|
|
miR-targeted genes in lymphocytes - TarBase
|
|
miR-targeted genes in epithelium - TarBase
|
|
Semaphorin interactions
|
| References |
| REF 1 | (http://www.guidetopharmacology.org/) Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5679). |
|---|
| REF 2 | Clinical pipeline report, company report or official report of Exelixis (2011). |
|---|
| REF 3 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. |
|---|