| Drug General Information |
| Drug ID |
D0N0TS
|
| Former ID |
DNC003673
|
| Drug Name |
PSAMMAPLIN A
|
| Drug Type |
Small molecular drug
|
| Indication |
Discovery agent
|
Investigative |
[1]
|
|---|
| Structure |
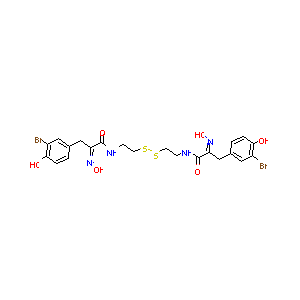
|
Download
2D MOL
3D MOL
|
| Formula |
C22H24Br2N4O6S2
|
| Canonical SMILES |
C1=CC(=C(C=C1CC(=NO)C(=O)NCCSSCCNC(=O)C(=NO)CC2=CC(=C(C<br />=C2)O)Br)Br)O
|
| InChI |
1S/C22H24Br2N4O6S2/c23-15-9-13(1-3-19(15)29)11-17(27-33)21(31)25-5-7-35-36-8-6-26-22(32)18(28-34)12-14-2-4-20(30)16(24)10-14/h1-4,9-10,29-30,33-34H,5-8,11-12H2,(H,25,31)(H,26,32)/b27-17-,28-18-
|
| InChIKey |
LMAFSGDNHVBIHU-HJTNQMAYSA-N
|
| PubChem Compound ID |
|
| Target and Pathway |
| Target(s) |
Histone Deacetylase 10 |
Target Info |
Inhibitor |
[2]
|
|---|
| Histone deacetylase 2 |
Target Info |
Inhibitor |
[2]
|
| Histone Deacetylase 8 |
Target Info |
Inhibitor |
[2]
|
| UDP-glucose 4-epimerase |
Target Info |
Inhibitor |
[1]
|
| Histone deacetylase |
Target Info |
Inhibitor |
[2]
|
| Histone deacetylase 6 |
Target Info |
Inhibitor |
[2]
|
| Histone deacetylase 1 |
Target Info |
Inhibitor |
[2]
|
| Histone deacetylase 4 |
Target Info |
Inhibitor |
[2]
|
|
BioCyc Pathway
|
D-galactose degradation V (Leloir pathway)
|
|
UDP-N-acetyl-D-galactosamine biosynthesis I
|
|
UDP-N-acetyl-D-galactosamine biosynthesis II
|
|
KEGG Pathway
|
Cell cycle
|
|
Notch signaling pathway
|
|
Thyroid hormone signaling pathway
|
|
Huntington's disease
|
|
Alcoholism
|
|
Epstein-Barr virus infection
|
|
Pathways in cancer
|
|
Transcriptional misregulation in cancer
|
|
Viral carcinogenesis
|
|
Chronic myeloid leukemiahsa05034:Alcoholism
|
|
Viral carcinogenesishsa00052:Galactose metabolism
|
|
Amino sugar and nucleotide sugar metabolism
|
|
Metabolic pathwayshsa05034:Alcoholism
|
|
Viral carcinogenesishsa04110:Cell cycle
|
|
Amphetamine addiction
|
|
MicroRNAs in cancer
|
|
NetPath Pathway
|
TCR Signaling PathwayNetPath_14:IL2 Signaling PathwayNetPath_11:TCR Signaling PathwayNetPath_14:IL2 Signaling Pathway
|
|
PANTHER Pathway
|
Wnt signaling pathwayP00057:Wnt signaling pathwayP02744:Fructose galactose metabolismP00057:Wnt signaling pathway
|
|
p53 pathway
|
|
Pathway Interaction Database
|
Signaling events mediated by HDAC Class II
|
|
Signaling events mediated by HDAC Class Ismad2_3nuclearpathway:Regulation of nuclear SMAD2/3 signaling
|
|
Signaling events mediated by HDAC Class I
|
|
Regulation of Telomerase
|
|
Glucocorticoid receptor regulatory network
|
|
Direct p53 effectors
|
|
Hedgehog signaling events mediated by Gli proteins
|
|
Regulation of nuclear beta catenin signaling and target gene transcriptionhdac_classi_pathway:Signaling events mediated by HDAC Class Ihdac_classii_pathway:Signaling events mediated by HDAC Class II
|
|
Notch signaling pathway
|
|
E2F transcription factor network
|
|
Presenilin action in Notch and Wnt signaling
|
|
Sumoylation by RanBP2 regulates transcriptional repression
|
|
Regulation of Androgen receptor activity
|
|
IL3-mediated signaling events
|
|
Validated nuclear estrogen receptor alpha network
|
|
Retinoic acid receptors-mediated signaling
|
|
Regulation of nuclear beta catenin signaling and target gene transcription
|
|
Validated targets of C-MYC transcriptional repression
|
|
Regulation of retinoblastoma protein
|
|
Notch-mediated HES/HEY networkhdac_classii_pathway:Signaling events mediated by HDAC Class II
|
|
Signaling events mediated by HDAC Class III
|
|
PathWhiz Pathway
|
Nucleotide Sugars Metabolism
|
|
Galactose Metabolism
|
|
Reactome
|
p75NTR negatively regulates cell cycle via SC1
|
|
NOTCH1 Intracellular Domain Regulates Transcription
|
|
Constitutive Signaling by NOTCH1 PEST Domain Mutants
|
|
Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants
|
|
HDACs deacetylate histones
|
|
NoRC negatively regulates rRNA expression
|
|
RNA Polymerase I Transcription Initiation
|
|
Factors involved in megakaryocyte development and platelet productionR-HSA-2122947:NOTCH1 Intracellular Domain Regulates Transcription
|
|
HDACs deacetylate histonesR-HSA-2122947:NOTCH1 Intracellular Domain Regulates Transcription
|
|
Assembly of the primary ciliumR-HSA-1538133:G0 and Early G1
|
|
Formation of the beta-catenin:TCF transactivating complex
|
|
SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription
|
|
Deactivation of the beta-catenin transactivating complex
|
|
WikiPathways
|
Notch Signaling Pathway
|
|
Notch Signaling Pathway
|
|
Neural Crest Differentiation
|
|
Signalling by NGF
|
|
RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription
|
|
Factors involved in megakaryocyte development and platelet production
|
|
Cell CycleWP2377:Integrated Pancreatic Cancer Pathway
|
|
Cell CycleWP1848:Metabolism of carbohydratesWP2858:Ectoderm Differentiation
|
|
Cell CycleWP706:SIDS Susceptibility Pathways
|
|
TGF beta Signaling Pathway
|
|
IL-6 signaling pathway
|
|
Apoptosis-related network due to altered Notch3 in ovarian cancer
|
|
Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer
|
|
Retinoblastoma (RB) in Cancer
|
|
TWEAK Signaling Pathway
|
|
Integrated Breast Cancer Pathway
|
|
Mitotic G1-G1/S phases
|
|
Cell Cycle
|
|
Androgen receptor signaling pathwayWP474:Endochondral Ossification
|
|
Cardiac Hypertrophic Response
|
|
miR-targeted genes in muscle cell - TarBase
|
|
miR-targeted genes in lymphocytes - TarBase
|
|
miR-targeted genes in epithelium - TarBase
|
|
TarBasePathway
|
|
MicroRNAs in cardiomyocyte hypertrophy
|
| References |
| REF 1 | Bioorg Med Chem Lett. 2006 Nov 15;16(22):5744-7. Epub 2006 Sep 7.Identification of novel inhibitors of UDP-Glc 4'-epimerase, a validated drug target for african sleeping sickness. |
|---|
| REF 2 | J Med Chem. 2003 Nov 20;46(24):5097-116.Histone deacetylase inhibitors. |
|---|