Drug Information
| Drug General Information | |||||
|---|---|---|---|---|---|
| Drug ID |
D08YAF
|
||||
| Former ID |
DNC006638
|
||||
| Drug Name |
PLSQETFSDLWKLLPEN-NH2
|
||||
| Indication | Discovery agent | Investigative | [528232] | ||
| Structure |
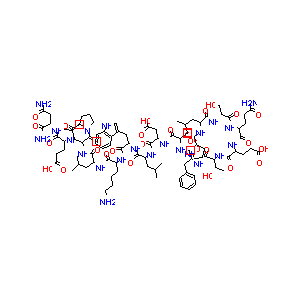
|
Download2D MOL |
|||
| Formula |
C93H142N22O28
|
||||
| Canonical SMILES |
CC(C)CC(C(=O)NC(CO)C(=O)NC(CCC(=O)N)C(=O)NC(CCC(=O)O)C(<br />=O)NC(C(C)O)C(=O)NC(CC1=CC=CC=C1)C(=O)NC(CO)C(=O)NC(CC(<br />=O)O)C(=O)NC(CC(C)C)C(=O)NC(CC2=CNC3=CC=CC=C32)C(=O)NC(<br />CCCCN)C(=O)NC(CC(C)C)C(=O)NC(CC(C)C)C(=O)N4CCCC4C(=O)NC<br />(CCC(=O)O)C(=O)NC(CC(=O)N)C(=O)N)NC(=O)C5CCCN5
|
||||
| InChI |
1S/C93H142N22O28/c1-46(2)35-61(83(133)108-65(40-52-43-99-54-22-14-13-21-53(52)54)87(137)100-56(23-15-16-32-94)79(129)106-62(36-47(3)4)84(134)111-67(38-49(7)8)93(143)115-34-18-25-70(115)91(141)103-58(27-30-73(121)122)81(131)104-60(77(97)127)41-72(96)120)107-88(138)66(42-75(125)126)109-90(140)69(45-117)113-86(136)64(39-51-19-11-10-12-20-51)110-92(142)76(50(9)118)114-82(132)59(28-31-74(123)124)101-80(130)57(26-29-71(95)119)102-89(139)68(44-116)112-85(135)63(37-48(5)6)105-78(128)55-24-17-33-98-55/h10-14,19-22,43,46-50,55-70,76,98-99,116-118H,15-18,23-42,44-45,94H2,1-9H3,(H2,95,119)(H2,96,120)(H2,97,127)(H,100,137)(H,101,130)(H,102,139)(H,103,141)(H,104,131)(H,105,128)(H,106,129)(H,107,138)(H,108,133)(H,109,140)(H,110,142)(H,111,134)(H,112,135)(H,113,136)(H,114,132)(H,121,122)(H,123,124)(H,125,126)/t50-,55-,56-,57-,58-,59-,60-,61-,62-,63-,64-,65-,66-,67-,68-,69-,70-,76-/m0/s1
|
||||
| InChIKey |
JGQYEMDYGMNCJX-RDLTZJDDSA-N
|
||||
| PubChem Compound ID | |||||
| Target and Pathway | |||||
| Target(s) | mRNA of Human mdm2 | Target Info | Inhibitor | [528232] | |
| KEGG Pathway | FoxO signaling pathway | ||||
| Cell cycle | |||||
| p53 signaling pathway | |||||
| Ubiquitin mediated proteolysis | |||||
| Endocytosis | |||||
| PI3K-Akt signaling pathway | |||||
| Thyroid hormone signaling pathway | |||||
| Epstein-Barr virus infection | |||||
| Pathways in cancer | |||||
| Transcriptional misregulation in cancer | |||||
| Viral carcinogenesis | |||||
| Proteoglycans in cancer | |||||
| MicroRNAs in cancer | |||||
| Glioma | |||||
| Prostate cancer | |||||
| Melanoma | |||||
| Bladder cancer | |||||
| Chronic myeloid leukemia | |||||
| Pathway Interaction Database | ErbB4 signaling events | ||||
| p73 transcription factor network | |||||
| ATR signaling pathway | |||||
| ATM pathway | |||||
| Glucocorticoid receptor regulatory network | |||||
| Sumoylation by RanBP2 regulates transcriptional repression | |||||
| Direct p53 effectors | |||||
| Regulation of Androgen receptor activity | |||||
| Validated transcriptional targets of deltaNp63 isoforms | |||||
| Aurora A signaling | |||||
| Validated transcriptional targets of TAp63 isoforms | |||||
| p53 pathway | |||||
| Regulation of retinoblastoma protein | |||||
| WikiPathways | DNA Damage Response (only ATM dependent) | ||||
| DNA Damage Response | |||||
| Senescence and Autophagy in Cancer | |||||
| Tryptophan metabolism | |||||
| G1 to S cell cycle control | |||||
| Copper homeostasis | |||||
| Bladder Cancer | |||||
| Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | |||||
| PIP3 activates AKT signaling | |||||
| Apoptosis | |||||
| ATM Signaling Pathway | |||||
| Retinoblastoma (RB) in Cancer | |||||
| Integrated Pancreatic Cancer Pathway | |||||
| Prostate Cancer | |||||
| Signaling Pathways in Glioblastoma | |||||
| Integrated Breast Cancer Pathway | |||||
| Integrated Cancer pathway | |||||
| Cell Cycle | |||||
| Cell Cycle Checkpoints | |||||
| TP53 Network | |||||
| miRNA Regulation of DNA Damage Response | |||||
| Androgen receptor signaling pathway | |||||
| References | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

