| Drug General Information |
| Drug ID |
D09XQF
|
| Drug Name |
Isoniazid |
|
| Synonyms |
Abdizide; Andrazide; Anidrasona; Antimicina; Antituberkulosum; Armacide; Armazid; Armazide; Atcotibine; Azuren; Bacillen; Bacillin; Cedin; Cemidon; Chemiazid; Chemidon; Continazine; Cortinazine; Cotinazin; Cotinizin; Defonin; Dibutin; Diforin; Dinacrin; Ditubin; Ebidene; Eralon; Ertuban; Eutizon; Evalon; Fetefu; Fimalene; HIA; Hidranizil; Hidrasonil; Hidrulta; Hidrun; Hycozid; Hydra; Hydrazid; Hydrazide; Hyozid; Hyzyd; INH; Idrazil; Inah; Inizid; Iscotin; Isidrina; Ismazide; Isobicina; Isocid; Isocidene; Isocotin; Isohydrazide; Isokin; Isolyn; Isonerit; Isonex; Isoniacid; Isoniazida; Isoniazide; Isoniazidum; Isonicazide; Isonicid; Isonico; Isonicotan; Isonicotil; Isonicotinhydrazid; Isonicotinohydrazide; Isonide; Isonidrin; Isonikazid; Isonilex; Isonin; Isonindon; Isonirit; Isoniton; Isonizida; Isonizide; Isotamine; Isotebe; Isotebezid; Isotinyl; Isozid; Isozide; Isozyd; LANIZID; Laniazid; Laniozid; Mybasan; Neoteben; Neoxin; Neumandin; Nevin; Niadrin; Nicazide; Nicetal; Nicizina; Niconyl; Nicotibina; Nicotibine; Nicotisan; Nicozide; Nidaton; Nidrazid; Nikozid; Niplen; Nitadon; Niteban; Nydrazid; Nyscozid; Pelazid; Percin; Phthisen; Pycazide; Pyreazid; Pyricidin; Pyridicin; Pyrizidin; Raumanon; Razide; Retozide; Rimicid; Rimifon; Rimiphone; Rimitsid; Robiselin; Robisellin; Roxifen; Sanohidrazina; Sauterazid; Sauterzid; Stanozide; Tebecid; Tebemid; Tebenic; Tebexin; Tebilon; Tebos; Teebaconin; Tekazin; Tibazide; Tibemid; Tibiazide; Tibinide; Tibison; Tibivis; Tibizide; Tibusan; Tisin; Tisiodrazida; Tizide; Tubazid; Tubazide; Tubeco; Tubecotubercid; Tubercid; Tuberian; Tubicon; Tubilysin; Tubizid; Tubomel; Tyvid; Unicocyde; Unicozyde; Vazadrine; Vederon; Zidafimia; Zinadon; Zonazide; Hid rasonil; Isoco tin; Isoniazid SA; Isozid e; Nidra zid; Rimif on; BP 5015; Bp 5 015; FSR 3; I0138; INHd20; L 1945; Nitebannsc 9659; Preparation 6424; RP 5015; AZT + Isoniazid; Cedin (Aerosol); Dow-Isoniazid; FRS-3; FSR-3; Ido-tebin; In-73; Inh-Burgthal; Isoniazid & EEP; Isoniazid & Propolis; Laniazid (TN); Neo-Tizide; Nydrazid (TN); RP-5015; TB-Phlogin; TB-Razide; TB-Vis; Usaf cb-2; I.A.I; RU-EF-Tb; RY-EF-Tb; I.A.I. |
| Drug Type |
Small molecular drug |
| Therapeutic Class |
Antitubercular Agents |
| Company |
Roche Pharmaceuticals |
| Structure |
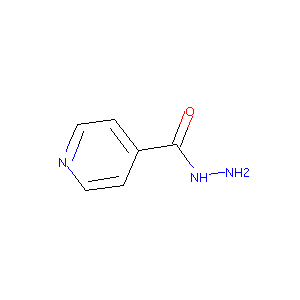
|
| Drug Resistance Mutations |
| Target Name |
Bacterial Catalase-peroxidase (katG) |
Target Info |
| Gene Name |
katG |
| Uniprot ID |
KATG_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MPEQHPPITETTTGAASNGCPVVGHMKYPVEGGGNQDWWPNRLNLKVLHQNPAVADPMGA
AFDYAAEVATIDVDALTRDIEEVMTTSQPWWPADYGHYGPLFIRMAWHAAGTYRIHDGRG
GAGGGMQRFAPLNSWPDNASLDKARRLLWPVKKKYGKKLSWADLIVFAGNCALESMGFKT
FGFGFGRVDQWEPDEVYWGKEATWLGDERYSGKRDLENPLAAVQMGLIYVNPEGPNGNPD
PMAAAVDIRETFRRMAMNDVETAALIVGGHTFGKTHGAGPADLVGPEPEAAPLEQMGLGW
KSSYGTGTGKDAITSGIEVVWTNTPTKWDNSFLEILYGYEWELTKSPAGAWQYTAKDGAG
AGTIPDPFGGPGRSPTMLATDLSLRVDPIYERITRRWLEHPEELADEFAKAWYKLIHRDM
GPVARYLGPLVPKQTLLWQDPVPAVSHDLVGEAEIASLKSQIRASGLTVSQLVSTAWAAA
SSFRGSDKRGGANGGRIRLQPQVGWEVNDPDGDLRKVIRTLEEIQESFNSAAPGNIKVSF
ADLVVLGGCAAIEKAAKAAGHNITVPFTPGRTDASQEQTDVESFAVLEPKADGFRNYLGK
GNPLPAEYMLLDKANLLTLSAPEMTVLVGGLRVLGANYKRLPLGVFTEASESLTNDFFVN
LLDMGITWEPSPADDGTYQGKDGSGKVKWTGSRVDLVFGSNSELRALVEVYGADDAQPKF
VQDFVAAWDKVMNLDRFDVR [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: S315T |
[1] |
| Mutation Frequency |
9 out of 34 isolates |
|
| Mutation info |
Missense: S315R |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: G593D |
[4], [6] |
|
| Mutation info |
Missense: H108E |
[4], [6] |
|
| Mutation info |
Frameshift: Plus 521 |
[6], [7] |
|
| Mutation info |
Missense: S140N |
[4], [6] |
|
| Mutation info |
Missense: S315G |
[4], [6] |
|
| Mutation info |
Missense: N138D |
[4], [9] |
|
| Mutation info |
Missense: N138S |
[4], [9] |
|
| Mutation info |
Missense: A110V |
[4], [11] |
|
| Mutation info |
Missense: A139P |
[4], [11] |
|
| Mutation info |
Missense: G279D |
[4], [12] |
|
| Mutation info |
Missense: Y229F |
[4], [13] |
|
| Mutation info |
Missense: D735N |
[4], [14] |
|
| Mutation info |
Missense: W107R |
[4], [14] |
|
| Mutation info |
Missense: W328C |
[16], [17] |
| Mutation Frequency |
1 out of 68 isolates |
|
| Mutation info |
Missense: G285D |
[4], [18] |
|
| Mutation info |
Missense: A350S |
[4], [19] |
|
| Mutation info |
Missense: D63E |
[4], [19] |
|
| Mutation info |
Missense: G629S |
[4], [19] |
|
| Mutation info |
Missense: H108Q |
[4], [19] |
|
| Mutation info |
Missense: I335T |
[4], [19] |
|
| Mutation info |
Missense: T262R |
[4], [19] |
|
| Mutation info |
Missense: W300G |
[4], [20] |
|
| Mutation info |
Missense: R104L |
[4], [21] |
|
| Mutation info |
Missense: A243S |
[4], [24] |
| Mutation Frequency |
2 out of 15 isolates |
|
| Mutation info |
Missense: A550D |
[4], [24] |
| Mutation Frequency |
2 out of 15 isolates |
|
| Mutation info |
Missense: D419H |
[4], [25] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Missense: G19D |
[4], [24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: G316D |
[2], [4] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: L587M |
[4], [26] |
| Mutation Frequency |
1 out of 39 isolates |
|
| Mutation info |
Missense: M126I |
[4], [24] |
| Mutation Frequency |
2 out of 15 isolates |
|
| Mutation info |
Missense: P2S |
[4], [24] |
| Mutation Frequency |
3 out of 15 isolates |
|
| Mutation info |
Missense: Q127P |
[4], [27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: Q224E |
[4], [24] |
| Mutation Frequency |
3 out of 15 isolates |
|
| Mutation info |
Missense: R463L |
[4], [26] |
| Mutation Frequency |
7 out of 39 isolates |
|
| Mutation info |
Missense: S17N |
[4], [24] |
| Mutation Frequency |
2 out of 15 isolates |
|
| Mutation info |
Missense: S315I |
[4], [17] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Missense: S315N |
[4], [17] |
| Mutation Frequency |
5 out of 28 isolates |
|
| Mutation info |
Missense: S457I |
[4], [28] |
| Mutation Frequency |
1 out of 154 isolates |
|
| Mutation info |
Missense: T275P |
[4], [26] |
| Mutation Frequency |
1 out of 39 isolates |
|
| Mutation info |
Missense: W328G |
[1], [4] |
| Mutation Frequency |
1 out of 34 isolates |
|
| Mutation info |
Missense: W91R |
[4], [29] |
| Mutation Frequency |
29 out of 38 isolates in all katG mutations |
|
| Mutation info |
Missense: Y337C |
[4], [24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: A291P |
[32] |
|
| Mutation info |
Missense: R128P |
[32] |
|
| Mutation info |
Missense: L336R |
[3] |
|
| Mutation info |
Nonsense: W300 STOP |
[3] |
|
| Mutation info |
Missense: A574V |
[6] |
|
| Mutation info |
Missense: D142A |
[6] |
|
| Mutation info |
Missense: D94A |
[6] |
|
| Mutation info |
Missense: F252L |
[6] |
|
| Mutation info |
Missense: F567S |
[6] |
|
| Mutation info |
Missense: G485V |
[6] |
|
| Mutation info |
Missense: G99E |
[6] |
|
| Mutation info |
Missense: I334T |
[6] |
|
| Mutation info |
Missense: I393N |
[6] |
|
| Mutation info |
Missense: I71N |
[6] |
|
| Mutation info |
Nonsense: K200 STOP |
[6] |
|
| Mutation info |
Missense: L148A |
[6] |
|
| Mutation info |
Missense: L150A |
[6] |
|
| Mutation info |
Frameshift: Plus 113 |
[6] |
|
| Mutation info |
Frameshift: Plus 120 |
[6] |
|
| Mutation info |
Frameshift: Plus 122 |
[6] |
|
| Mutation info |
Frameshift: Plus 123 |
[6] |
|
| Mutation info |
Frameshift: Plus 217 |
[6] |
|
| Mutation info |
Frameshift: Plus 289 |
[6] |
|
| Mutation info |
Frameshift: Plus 511 |
[6] |
|
| Mutation info |
Frameshift: Plus 513 |
[6] |
|
| Mutation info |
Missense: S140A |
[6] |
|
| Mutation info |
Missense: S160L |
[6] |
|
| Mutation info |
Missense: T180C |
[6] |
|
| Mutation info |
Missense: V710A |
[6] |
|
| Mutation info |
Nonsense: W198 STOP |
[6] |
|
| Mutation info |
Nonsense: W477 STOP |
[6] |
|
| Mutation info |
Missense: N138H |
[9] |
|
| Mutation info |
Missense: A61T |
[34] |
|
| Mutation info |
Nonsense: D37 Truncation |
[34] |
|
| Mutation info |
Missense: D73N |
[34] |
|
| Mutation info |
Nonsense: E10 Truncation |
[34] |
|
| Mutation info |
Nonsense: F62 Truncation |
[34] |
|
| Mutation info |
Nonsense: G33 Truncation |
[34] |
|
| Mutation info |
Missense: N35D |
[34] |
|
| Mutation info |
Missense: T11A |
[34] |
|
| Mutation info |
Missense: T12P |
[34] |
|
| Mutation info |
Missense: D735A |
[11] |
|
| Mutation info |
Missense: L619P |
[11] |
|
| Mutation info |
Missense: L634F |
[11] |
|
| Mutation info |
Missense: N529D |
[29] |
| Mutation Frequency |
29 out of 38 isolates in all katG mutations |
|
| Mutation info |
Frameshift: Plus 124 |
[29] |
|
| Mutation info |
Nonsense: Q434 STOP |
[29] |
|
| Mutation info |
Missense: G309D |
[36] |
|
| Mutation info |
Missense: E195K |
[12] |
|
| Mutation info |
Missense: N138T |
[12] |
|
| Mutation info |
Missense: W341S |
[12] |
|
| Mutation info |
Missense: A727D |
[37] |
|
| Mutation info |
Missense: G307E |
[37] |
|
| Mutation info |
Nonsense: P29 Truncation |
[37] |
|
| Mutation info |
Missense: R249C |
[37] |
|
| Mutation info |
Missense: T275S |
[37] |
|
| Mutation info |
Missense: R418Q |
[38] |
|
| Mutation info |
Missense: G234E |
[39] |
|
| Mutation info |
Missense: G699Q |
[39] |
|
| Mutation info |
Frameshift: Minus 1 |
[39] |
|
| Mutation info |
Missense: P241P |
[39] |
|
| Mutation info |
Frameshift: Plus 50 |
[39] |
|
| Mutation info |
Missense: R484S |
[39] |
|
| Mutation info |
Missense: R490C |
[39] |
|
| Mutation info |
Nonsense: W107 STOP |
[39] |
|
| Mutation info |
Missense: W191R |
[39] |
|
| Mutation info |
Missense: R418L |
[13] |
|
| Mutation info |
Missense: D117A |
[40] |
| Mutation Frequency |
52 out of 34 isolates in all katG mutations |
|
| Mutation info |
Missense: G491C |
[40] |
| Mutation Frequency |
52 out of 34 isolates in all katG mutations |
|
| Mutation info |
Missense: M257I |
[40] |
| Mutation Frequency |
52 out of 34 isolates in all katG mutations |
|
| Mutation info |
Missense: G685R |
[7] |
|
| Mutation info |
Frameshift: Plus 438 |
[7] |
|
| Mutation info |
Missense: R515C |
[7] |
|
| Mutation info |
Missense: A106V |
[14] |
| Mutation Frequency |
128 out of 143 isolates in all katG mutations |
|
| Mutation info |
Missense: A109V |
[14] |
|
| Mutation info |
Missense: A162T |
[14] |
|
| Mutation info |
Missense: A636E |
[14] |
|
| Mutation info |
Missense: D573N |
[14] |
|
| Mutation info |
Deletion: D735 |
[14] |
|
| Mutation info |
Missense: D94G |
[14] |
|
| Mutation info |
Missense: E607K |
[14] |
|
| Mutation info |
Missense: G121C |
[14] |
|
| Mutation info |
Missense: K143T |
[14] |
|
| Mutation info |
Missense: K345T |
[14] |
|
| Mutation info |
Missense: L384R |
[14] |
|
| Mutation info |
Missense: L653P |
[14] |
|
| Mutation info |
Missense: P280H |
[14] |
|
| Mutation info |
Frameshift: Plus 301 |
[14] |
|
| Mutation info |
Missense: R104Q |
[14] |
|
| Mutation info |
Missense: T251M |
[14] |
|
| Mutation info |
Missense: T394A |
[14] |
|
| Mutation info |
Missense: T85P |
[14] |
|
| Mutation info |
Missense: V1L |
[14] |
|
| Mutation info |
Missense: W728C |
[14] |
|
| Mutation info |
Missense: Y155C |
[14] |
| Mutation Frequency |
69 out of 101 isolates in all embB mutations |
|
| Mutation info |
Missense: A172T |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: A574E |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: A713P |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: D72K |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: L587I |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Frameshift: Plus 556 |
[2] |
| Mutation Frequency |
3 out of 238 patients in all katG mutations |
|
| Mutation info |
Missense: Q717P |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: S140G |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: T180K |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Nonsense: W38 STOP |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Nonsense: W397 STOP |
[2] |
| Mutation Frequency |
3 out of 238 isolates in all katG mutations |
|
| Mutation info |
Missense: A172V |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: A409D |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: D311G |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: D72G |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: D74G |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: D74Y |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: E289D |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: G125C |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: G186H |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: G186V |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: G234R |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: G299C |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: G96C |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: I317L |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: M176I |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: M84I |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: N218K |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: N236T |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Frameshift: Plus 191 |
[27] |
|
| Mutation info |
Frameshift: Plus 268 |
[27] |
|
| Mutation info |
Frameshift: Plus 346 |
[27] |
| Mutation Frequency |
38 out of 47 patients in all katG mutations |
|
| Mutation info |
Missense: T380I |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: T390I |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: V230A |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: W321R |
[27] |
| Mutation Frequency |
38 out of 47 isolates in all katG mutations |
|
| Mutation info |
Missense: I335V |
[42] |
| Mutation Frequency |
207 out of 217 isolates |
|
| Mutation info |
Missense: T308P |
[43] |
|
| Mutation info |
Missense: G297V |
[44] |
|
| Mutation info |
Missense: L141F |
[44] |
|
| Mutation info |
Insertion: L390 |
[44] |
|
| Mutation info |
Missense: L48Q |
[44] |
|
| Mutation info |
Missense: M257T |
[44] |
|
| Mutation info |
Missense: Q295P |
[44] |
|
| Mutation info |
Missense: D311F |
[16] |
|
| Mutation info |
Missense: D311Y |
[16] |
|
| Mutation info |
Missense: D357H |
[16] |
| Mutation Frequency |
2 out of 163 isolates |
|
| Mutation info |
Missense: D357N |
[16] |
| Mutation Frequency |
2 out of 163 isolates |
|
| Mutation info |
Missense: E454R |
[16] |
|
| Mutation info |
Missense: G305A |
[16] |
|
| Mutation info |
Missense: G307A |
[16] |
|
| Mutation info |
Missense: G307R |
[16] |
|
| Mutation info |
Missense: G309C |
[16] |
|
| Mutation info |
Missense: G309F |
[16] |
|
| Mutation info |
Missense: G309S |
[16] |
|
| Mutation info |
Missense: G316S |
[16] |
|
| Mutation info |
Missense: S314N |
[16] |
| Mutation Frequency |
3 out of 163 isolates |
|
| Mutation info |
Missense: S314T |
[16] |
|
| Mutation info |
Nonsense: W321 STOP |
[16] |
|
| Mutation info |
Missense: W321C |
[16] |
|
| Mutation info |
Missense: W321L |
[16] |
|
| Mutation info |
Missense: W321S |
[16] |
|
| Mutation info |
Missense: W204R |
[18] |
|
| Mutation info |
Missense: W505S |
[18] |
|
| Mutation info |
Missense: L449F |
[46] |
| Mutation Frequency |
1 out of 48 isolates |
|
| Mutation info |
Nonsense: Q352 STOP |
[47] |
| Mutation Frequency |
1 out of 110 isolates |
|
| Mutation info |
Missense: W328S |
[47] |
| Mutation Frequency |
3 out of 24 isolates |
|
| Mutation info |
Missense: A361D |
[25] |
|
| Mutation info |
Missense: A379V |
[25] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Missense: G299A |
[25] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Missense: L378P |
[25] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Frameshift: Plus 310 |
[25] |
|
| Mutation info |
Frameshift: Plus 364 |
[25] |
|
| Mutation info |
Frameshift: Plus 401 |
[25] |
|
| Mutation info |
Missense: Y413H |
[25] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Missense: K414N |
[28] |
| Mutation Frequency |
1 out of 154 isolates |
|
| Mutation info |
Missense: Y304S |
[28] |
| Mutation Frequency |
1 out of 154 isolates |
|
| Mutation info |
Missense: D311E |
[48] |
| Mutation Frequency |
409 out of 526 isolates |
|
| Mutation info |
Missense: G318V |
[48] |
|
| Mutation info |
Missense: S331C |
[48] |
|
| Mutation info |
Missense: A444T |
[] |
|
| Mutation info |
Missense: A551G |
[] |
|
| Mutation info |
Missense: I462T |
[] |
|
| Mutation info |
Missense: N238K |
[] |
|
| Mutation info |
Missense: Q502H |
[] |
|
| Mutation info |
Missense: S140R |
[] |
|
| Mutation info |
Deletion: G120-G123 |
[26] |
| Mutation Frequency |
2 out of 39 isolates |
|
| Mutation info |
Insertion: H125 |
[26] |
| Mutation Frequency |
1 out of 39 isolates |
|
| Mutation info |
Missense: V1A |
[26] |
| Mutation Frequency |
1 out of 39 isolates |
|
| Mutation info |
Missense: A264T |
[24] |
| Mutation Frequency |
2 out of 15 isolates |
|
| Mutation info |
Missense: A281V |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: A424E |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: A424V |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: A65T |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: A66P |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: G169A |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: M609I |
[24] |
| Mutation Frequency |
3 out of 15 isolates |
|
| Mutation info |
Missense: N258S |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: P429S |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: P589T |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Frameshift: Plus 10 |
[24] |
| Mutation Frequency |
1 out of 15 patients |
|
| Mutation info |
Missense: S302R |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: W505R |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Nonsense: W90 STOP |
[24] |
| Mutation Frequency |
1 out of 15 isolates |
|
| Mutation info |
Missense: Y337F |
[24] |
| Mutation Frequency |
2 out of 15 isolates |
|
| Mutation info |
Missense: A716P |
[19] |
|
| Mutation info |
Missense: A409R |
[49] |
|
| Mutation info |
Deletion: A52-P57 |
[49] |
|
| Mutation info |
Missense: D695A |
[49] |
|
| Mutation info |
Missense: T275A |
[49] |
|
| Mutation info |
Missense: K488N |
[20] |
|
| Mutation info |
Missense: L587P |
[20] |
|
| Mutation info |
Frameshift: Plus 617 |
[20] |
|
| Mutation info |
Frameshift: Plus 65 |
[20] |
|
| Mutation info |
Missense: Q525P |
[20] |
|
| Mutation info |
Missense: S315L |
[20] |
|
| Mutation info |
Missense: S700P |
[20] |
|
| Mutation info |
Missense: A350T |
[21] |
|
| Mutation info |
Missense: D381G |
[21] |
|
| Mutation info |
Missense: H108D |
[21] |
|
| Mutation info |
Missense: L148R |
[21] |
|
| Mutation info |
Missense: W328L |
[17] |
| Mutation Frequency |
1 out of 68 isolates |
|
| Mutation info |
Missense: G121V |
[51] |
|
| Mutation info |
Missense: Q88R |
[51] |
| Mutation Frequency |
24 out of 27 isolates in all katG mutations |
|
| Mutation info |
Missense: Y155S |
[51] |
|
| Mutation info |
Missense: A139V |
[4] |
|
| Mutation info |
Missense: A162V |
[4] |
|
| Mutation info |
Missense: A256T |
[4] |
|
| Mutation info |
Missense: A379T |
[4] |
|
| Mutation info |
Missense: A541D |
[4] |
|
| Mutation info |
Missense: A93T |
[4] |
|
| Mutation info |
Missense: D142G |
[4] |
|
| Mutation info |
Missense: D194G |
[4] |
|
| Mutation info |
Missense: D194Y |
[4] |
|
| Mutation info |
Missense: D387G |
[4] |
|
| Mutation info |
Missense: D387H |
[4] |
|
| Mutation info |
Missense: D542H |
[4] |
|
| Mutation info |
Missense: F483L |
[4] |
|
| Mutation info |
Missense: G123E |
[4] |
|
| Mutation info |
Missense: G269D |
[4] |
|
| Mutation info |
Missense: G269R |
[4] |
|
| Mutation info |
Missense: G299S |
[4] |
|
| Mutation info |
Missense: G32D |
[4] |
|
| Mutation info |
Missense: H276M |
[4] |
|
| Mutation info |
Missense: H97R |
[4] |
|
| Mutation info |
Missense: L101P |
[4] |
|
| Mutation info |
Missense: M176T |
[4] |
|
| Mutation info |
Missense: M420T |
[4] |
|
| Mutation info |
Missense: M624V |
[4] |
|
| Mutation info |
Missense: N133T |
[4] |
|
| Mutation info |
Missense: P131Q |
[4] |
|
| Mutation info |
Missense: P131R |
[4] |
|
| Mutation info |
Missense: P131T |
[4] |
|
| Mutation info |
Missense: P232S |
[4] |
|
| Mutation info |
Missense: Q127E |
[4] |
|
| Mutation info |
Missense: Q295H |
[4] |
|
| Mutation info |
Missense: Q352E |
[4] |
|
| Mutation info |
Missense: Q439P |
[4] |
|
| Mutation info |
Missense: R128Q |
[4] |
|
| Mutation info |
Missense: R385W |
[4] |
|
| Mutation info |
Missense: R489S |
[4] |
|
| Mutation info |
Missense: R632C |
[4] |
|
| Mutation info |
Missense: S17T |
[4] |
|
| Mutation info |
Missense: S383P |
[4] |
|
| Mutation info |
Missense: T306P |
[4] |
|
| Mutation info |
Missense: T324P |
[4] |
|
| Mutation info |
Missense: T394M |
[4] |
|
| Mutation info |
Missense: T667P |
[4] |
|
| Mutation info |
Missense: V47E |
[4] |
|
| Mutation info |
Missense: V68G |
[4] |
|
| Mutation info |
Missense: Y64S |
[4] |
|
| Mutation info |
Missense: Y95C |
[4] |
|
| Mutation info |
Missense: Y98C |
[4] |
|
| Target Name |
Bacterial 3-oxoacyl-[acyl-carrier-protein] synthase 1 (kasA) |
Target Info |
| Gene Name |
kasA |
| Uniprot ID |
FAB1_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MSQPSTANGGFPSVVVTAVTATTSISPDIESTWKGLLAGESGIHALEDEFVTKWDLAVKI
GGHLKDPVDSHMGRLDMRRMSYVQRMGKLLGGQLWESAGSPEVDPDRFAVVVGTGLGGAE
RIVESYDLMNAGGPRKVSPLAVQMIMPNGAAAVIGLQLGARAGVMTPVSACSSGSEAIAH
AWRQIVMGDADVAVCGGVEGPIEALPIAAFSMMRAMSTRNDEPERASRPFDKDRDGFVFG
EAGALMLIETEEHAKARGAKPLARLLGAGITSDAFHMVAPAADGVRAGRAMTRSLELAGL
SPADIDHVNAHGTATPIGDAAEANAIRVAGCDQAAVYAPKSALGHSIGAVGALESVLTVL
TLRDGVIPPTLNYETPDPEIDLDVVAGEPRYGDYRYAVNNSFGFGGHNVALAFGRY [My
cobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: G387D |
[3], [4] |
|
| Mutation info |
Missense: R121K |
[3], [5] |
|
| Mutation info |
Missense: D66N |
[4], [22] |
|
| Mutation info |
Missense: F413L |
[4], [22] |
|
| Mutation info |
Missense: G269S |
[4], [22] |
|
| Mutation info |
Missense: G312S |
[4], [22] |
|
| Mutation info |
Missense: M77I |
[29] |
|
| Target Name |
Bacterial Enoyl-[acyl-carrier-protein] reductase [NADH] (Inha) |
Target Info |
| Gene Name |
inhA |
| Uniprot ID |
INHA_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTGLLDGKRILVSGIITDSSIAFHIARVAQEQGAQLVLTGFDRLRLIQRITDRLPAKAPL
LELDVQNEEHLASLAGRVTEAIGAGNKLDGVVHSIGFMPQTGMGINPFFDAPYADVSKGI
HISAYSYASMAKALLPIMNPGGSIVGMDFDPSRAMPAYNWMTVAKSALESVNRFVAREAG
KYGVRSNLVAAGPIRTLAMSAIVGGALGEEAGAQIQLLEEGWDQRAPIGWNMKDATPVAK
TVCALLSDWLPATTGDIIYADGGAHTQLL [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: S94A |
[5], [8] |
|
| Mutation info |
Missense: I21T |
[5], [12] |
|
| Mutation info |
Missense: I21V |
[5], [12] |
|
| Mutation info |
Missense: I16T |
[5], [23] |
|
| Mutation info |
Missense: I47T |
[5], [23] |
|
| Mutation info |
Missense: V78A |
[5], [23] |
|
| Mutation info |
Missense: I194T |
[5], [30] |
| Mutation Frequency |
52 out of 102 isolates |
|
| Mutation info |
Missense: I95P |
[5], [31] |
| Mutation Frequency |
7 out of 14 isolates |
|
| Mutation info |
Missense: I95T |
[6] |
|
| Mutation info |
Missense: S94L |
[41] |
|
| Mutation info |
Missense: E217D |
[25] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Missense: R202G |
[25] |
| Mutation Frequency |
1 out of 28 isolates |
|
| Mutation info |
Missense: K8N |
[] |
|
| Target Name |
Bacterial Probable NADH dehydrogenase (Ndh) |
Target Info |
| Gene Name |
ndh |
| Uniprot ID |
P95160_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MSPQQEPTAQPPRRHRVVIIGSGFGGLNAAKKLKRADVDIKLIARTTHHLFQPLLYQVAT
GIISEGEIAPPTRVVLRKQRNVQVLLGNVTHIDLAGQCVVSELLGHTYQTPYDSLIVAAG
AGQSYFGNDHFAEFAPGMKSIDDALELRGRILSAFEQAERSSDPERRAKLLTFTVVGAGP
TGVEMAGQIAELAEHTLKGAFRHIDSTKARVILLDAAPAVLPPMGAKLGQRAAARLQKLG
VEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSAGVSASRLGRDLAEQSRVELDRA
GRVQVLPDLSIPGYPNVFVVGDMAAVEGVPGVAQGAIQGAKYVASTIKAELAGANPAERE
PFQYFDKGSMATVSRFSAVAKIGPVEFSGFIAWLIWLVLHLAYLIGFKTKITTLLSWTVT
FLSTRRGQLTITDQQAFARTRLEQLAELAAEAQGSAASAKVAS [Mycobacterium t
uberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: R268H |
[4], [10] |
|
| Mutation info |
Missense: T110A |
[4], [10] |
|
| Mutation info |
Missense: R13C |
[4], [15] |
|
| Mutation info |
Missense: V18A |
[4], [15] |
|
| Mutation info |
Missense: L239P |
[2] |
| Mutation Frequency |
100 out of 144 isolates in all ndh mutations |
|
| Mutation info |
Missense: A300P |
[4] |
|
| Mutation info |
Missense: G313R |
[4] |
|
| Mutation info |
Missense: L50V |
[4] |
|
| Target Name |
Bacterial Isoniazid-induced protein (IniA) |
Target Info |
| Gene Name |
inia |
| Uniprot ID |
INIA_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MVPAGLCAYRDLRRKRARKWGDTVTQPDDPRRVGVIVELIDHTIAIAKLNERGDLVQRLT
RARQRITDPQVRVVIAGLLKQGKSQLLNSLLNLPAARVGDDEATVVITVVSYSAQPSARL
VLAAGPDGTTAAVDIPVDDISTDVRRAPHAGGREVLRVEVGAPSPLLRGGLAFIDTPGVG
GLGQPHLSATLGLLPEADAVLVVSDTSQEFTEPEMWFVRQAHQICPVGAVVATKTDLYPR
WREIVNANAAHLQRARVPMPIIAVSSLLRSHAVTLNDKELNEESNFPAIVKFLSEQVLSR
ATERVRAGVLGEIRSATEQLAVSLGSELSVVNDPNLRDRLASDLERRKREAQQAVQQTAL
WQQVLGDGFNDLTADVDHDLRTRFRTVTEDAERQIDSCDPTAHWAEIGNDVENAIATAVG
DNFVWAYQRSEALADDVARSFADAGLDSVLSAELSPHVMGTDFGRLKALGRMESKPLRRG
HKMIIGMRGSYGGVVMIGMLSSVVGLGLFNPLSVGAGLILGRMAYKEDKQNRLLRVRSEA
KANVRRFVDDISFVVSKQSRDRLKMIQRLLRDHYREIAEEITRSLTESLQATIAAAQVAE
TERDNRIRELQRQLGILSQVNDNLAGLEPTLTPRASLGRA [Mycobacterium tube
rculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: P3A |
[29] |
|
| Mutation info |
Frameshift: Plus 94 |
[29] |
|
| Mutation info |
Missense: R537H |
[29] |
|
| Target Name |
Bacterial Thioredoxin peroxidase (ahpC) |
Target Info |
| Gene Name |
ahpC |
| Uniprot ID |
AHPC_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MPLLTIGDQFPAYQLTALIGGDLSKVDAKQPGDYFTTITSDEHPGKWRVVFFWPKDFTFV
CPTEIAAFSKLNDEFEDRDAQILGVSIDSEFAHFQWRAQHNDLKTLPFPMLSDIKRELSQ
AAGVLNADGVADRVTFIVDPNNEIQFVSATAGSVGRNVDEVLRVLDALQSDELCACNWRK
GDPTLDAGELLKASA [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: P2S |
[3] |
|
| Mutation info |
Missense: T5I |
[3] |
|
| Mutation info |
Missense: D33N |
[6] |
|
| Mutation info |
Missense: L3K |
[33] |
|
| Mutation info |
Missense: F10I |
[35] |
|
| Mutation info |
Missense: D73H |
[2] |
| Mutation Frequency |
19 out of 73 isolates in all ahpC mutations |
|
| Mutation info |
Missense: L191R |
[2] |
| Mutation Frequency |
19 out of 73 isolates in all ahpC mutations |
|
| Mutation info |
Missense: E76K |
[27] |
|
| Target Name |
Bacterial Arylamine N-acetyltransferase (nhoA) |
Target Info |
| Gene Name |
nhoA |
| Uniprot ID |
NAT_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MALDLTAYFDRINYRGATDPTLDVLQDLVTVHSRTIPFENLDPLLGVPVDDLSPQALADK
LVLRRRGGYCFEHNGLMGYVLAELGYRVRRFAARVVWKLAPDAPLPPQTHTLLGVTFPGS
GGCYLVDVGFGGQTPTSPLRLETGAVQPTTHEPYRLEDRVDGFVLQAMVRDTWQTLYEFT
TQTRPQIDLKVASWYASTHPASKFVTGLTAAVITDDARWNLSGRDLAVHRAGGTEKIRLA
DAAAVVDTLSERFGINVADIGERGALETRIDELLARQPGADAP [Mycobacterium t
uberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: G207E |
[29] |
| Mutation Frequency |
6 out of 38 isolates in all nhoA mutations |
|
| Mutation info |
Missense: G67R |
[29] |
| Mutation Frequency |
6 out of 38 isolates in all nhoA mutations |
|
| Target Name |
Bacterial Uncharacterized protein (Rv2242) |
Target Info |
| Gene Name |
Rv2242 |
| Uniprot ID |
Y2242_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MNDNQLAPVARPRSPLELLDTVPDSLLRRLKQYSGRLATEAVSAMQERLPFFADLEASQR
ASVALVVQTAVVNFVEWMHDPHSDVGYTAQAFELVPQDLTRRIALRQTVDMVRVTMEFFE
EVVPLLARSEEQLTALTVGILKYSRDLAFTAATAYADAAEARGTWDSRMEASVVDAVVRG
DTGPELLSRAAALNWDTTAPATVLVGTPAPGPNGSNSDGDSERASQDVRDTAARHGRAAL
TDVHGTWLVAIVSGQLSPTEKFLKDLLAAFADAPVVIGPTAPMLTAAHRSASEAISGMNA
VAGWRGAPRPVLARELLPERALMGDASAIVALHTDVMRPLADAGPTLIETLDAYLDCGGA
IEACARKLFVHPNTVRYRLKRITDFTGRDPTQPRDAYVLRVAATVGQLNYPTPH [Myco
bacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: D3G |
[29] |
|
| Mutation info |
Missense: M323T |
[29] |
|
| Mutation info |
Missense: T4A |
[29] |
|
| Target Name |
Bacterial Probable inactive lipase (Rv1592c) |
Target Info |
| Gene Name |
Rv1592c |
| Uniprot ID |
Y1592_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MVEPGNLAGATGAEWIGRPPHEELQRKVRPLLPSDDPFYFPPAGYQHAVPGTVLRSRDVE
LAFMGLIPQPVTATQLLYRTTNMYGNPEATVTTVIVPAELAPGQTCPLLSYQCAIDAMSS
RCFPSYALRRRAKALGSLTQMELLMISAALAEGWAVSVPDHEGPKGLWGSPYEPGYRVLD
GIRAALNSERVGLSPATPIGLWGYSGGGLASAWAAEACGEYAPDLDIVGAVLGSPVGDLG
HTFRRLNGTLLAGLPALVVAALQHSYPGLARVIKEHANDEGRQLLEQLTEMTTVDAVIRM
AGRDMGDFLDEPLEDILSTPEISHVFGDTKLGSAVPTPPVLIVQAVHDYLIDVSDIDALA
DSYTAGGANVTYHRDLFSEHVSLHPLSAPMTLRWLTDRFAGKPLTDHRVRTTWPTIFNPM
TYAGMARLAVIAAKVITGRKLSRRPL [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: P42L |
[29] |
|
| Mutation info |
Missense: V430A |
[29] |
|
| Target Name |
Bacterial Isoniazid-induced protein (IniC) |
Target Info |
| Gene Name |
iniC |
| Uniprot ID |
INIC_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MSTSDRVRAILHATIQAYRGAPAYRQRGDVFCQLDRIGARLAEPLRIALAGTLKAGKSTL
VNALVGDDIAPTDATEATRIVTWFRHGPTPRVTANHRGGRRANVPITRRGGLSFDLRRIN
PAELIDLEVEWPAEELIDATIVDTPGTSSLACDASERTLRLLVPADGVPRVDAVVFLLRT
LNAADVALLKQIGGLVGGSVGALGIIGVASRADEIGAGRIDAMLSANDVAKRFTRELNQM
GICQAVVPVSGLLALTARTLRQTEFIALRKLAGAERTELNRALLSVDRFVRRDSPLPVDA
GIRAQLLERFGMFGIRMSIAVLAAGVTDSTGLAAELLERSGLVALRNVIDQQFAQRSDML
KAHTALVSLRRFVQTHPVPATPYVIADIDPLLADTHAFEELRMLSLLPSRATTLNDDEIA
SLRRIIGGSGTSAAARLGLDPANSREAPRAALAAAQHWRRRAAHPLNDPFTTRACRAAVR
SAEAMVAEFSARR [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: W83G |
[29] |
|
| Target Name |
Bacterial Fibronectin-binding protein C (fbpC) |
Target Info |
| Gene Name |
fbpC |
| Uniprot ID |
A85C_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTFFEQVRRLRSAATTLPRRLAIAAMGAVLVYGLVGTFGGPATAGAFSRPGLPVEYLQVP
SASMGRDIKVQFQGGGPHAVYLLDGLRAQDDYNGWDINTPAFEEYYQSGLSVIMPVGGQS
SFYTDWYQPSQSNGQNYTYKWETFLTREMPAWLQANKGVSPTGNAAVGLSMSGGSALILA
AYYPQQFPYAASLSGFLNPSEGWWPTLIGLAMNDSGGYNANSMWGPSSDPAWKRNDPMVQ
IPRLVANNTRIWVYCGNGTPSDLGGDNIPAKFLEGLTLRTNQTFRDTYAADGGRNGVFNF
PPNGTHSWPYWNEQLVAMKADIQHVLNGATPPAAPAAPAA [Mycobacterium tube
rculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: G158S |
[29] |
|
| Target Name |
Bacterial Propanoyl-CoA:carbon dioxide ligase (accD6) |
Target Info |
| Gene Name |
accD6 |
| Uniprot ID |
PCC6_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTIMAPEAVGESLDPRDPLLRLSNFFDDGSVELLHERDRSGVLAAAGTVNGVRTIAFCTD
GTVMGGAMGVEGCTHIVNAYDTAIEDQSPIVGIWHSGGARLAEGVRALHAVGQVFEAMIR
ASGYIPQISVVVGFAAGGAAYGPALTDVVVMAPESRVFVTGPDVVRSVTGEDVDMASLGG
PETHHKKSGVCHIVADDELDAYDRGRRLVGLFCQQGHFDRSKAEAGDTDIHALLPESSRR
AYDVRPIVTAILDADTPFDEFQANWAPSMVVGLGRLSGRTVGVLANNPLRLGGCLNSESA
EKAARFVRLCDAFGIPLVVVVDVPGYLPGVDQEWGGVVRRGAKLLHAFGECTVPRVTLVT
RKTYGGAYIAMNSRSLNATKVFAWPDAEVAVMGAKAAVGILHKKKLAAAPEHEREALHDQ
LAAEHERIAGGVDSALDIGVVDEKIDPAHTRSKLTEALAQAPARRGRHKNIPL [Mycob
acterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: D229G |
[29] |
|
| Target Name |
Bacterial Efflux protein A (efpA) |
Target Info |
| Gene Name |
efpA |
| Uniprot ID |
EFPA_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTALNDTERAVRNWTAGRPHRPAPMRPPRSEETASERPSRYYPTWLPSRSFIAAVIAIGG
MQLLATMDSTVAIVALPKIQNELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRT
FIVGVALFTISSVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPARNAAT
AVFAAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMIYLARTALRETNKERMKLDA
TGAILATLACTAAVFAFSIGPEKGWMSGITIGSGLVALAAAVAFVIVERTAENPVVPFHL
FRDRNRLVTFSAILLAGGVMFSLTVCIGLYVQDILGYSALRAGVGFIPFVIAMGIGLGVS
SQLVSRFSPRVLTIGGGYLLFGAMLYGSFFMHRGVPYFPNLVMPIVVGGIGIGMAVVPLT
LSAIAGVGFDQIGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDVQL
AALDHAYTYGLLWVAGAAIIVGGMALFIGYTPQQVAHAQEVKEAIDAGEL [Mycobact
erium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: I73T |
[29] |
|
| Target Name |
Bacterial Conserved protein (Rv0340) |
Target Info |
| Gene Name |
Rv0340 |
| Uniprot ID |
O33270_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MANSLLDFVISLVRDPEAAARYAANPERSIAEAHLTDVTRADVNSLIPVVSDSLSMSEPI
GAAGGAHAGDRGNVWASGAATAALDAFAPHADAGVVQQHGAVGSVLNQPTPPGPGVTPTD
PRPFRAGPHETSALLTSAEIPDTTSEDGGLPTDHPAVWNHPVVDPHTVEPDHHGYDIHG
[Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: V163I |
[29] |
|
| Target Name |
Bacterial Probable acyl-CoA dehydrogenase (FadE24) |
Target Info |
| Gene Name |
fadE24 |
| Uniprot ID |
P95187_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTNTTSAANAAKPSGARTDRRGRTTGVGLAPHKRTGIDVALALLTPIVGQEFLDKYRLRD
PLNRSLRYGVKTMFATAGAATRQFQRVQGLRGGPTRLKSSGRDYFDLTPDDDQKLIIETV
DEFAEEVLRPAAHDADDAATYPSDLTAKAAELGITAINIPEDFDGIAEHRSSVTNVLVAE
ALAYGDMGLALPILAPGGVASALTHWGSADQQATYLKEFAGENVPQACVAITEPQPLFDP
TRLKTTAVRTPSGYRLDGVKSLIPAAADAELFIVGAQLGGKPALFIVESAASGLTVKADP
SMGIRGAALGQVELCGVSVPLNARLGEDEASDNDYSEALALARLGWAALAVGTSHAVLDY
VVPYVKQRQAFGEPIAHRQAVAFMCANIAIELDGLRLITWRGASRAEQGLPFAREAALAK
RLGSDKGMQIGLDGVQLLGGHGYTKEHPVERWYRDLRAIGVAEGVVVI [Mycobacter
ium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Frameshift: Minus 64 |
[29] |
|
| Target Name |
Bacterial Malonyl CoA-acyl carrier protein transacylase (fabD) |
Target Info |
| Gene Name |
fabD |
| Uniprot ID |
FABD_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MIALLAPGQGSQTEGMLSPWLQLPGAADQIAAWSKAADLDLARLGTTASTEEITDTAVAQ
PLIVAATLLAHQELARRCVLAGKDVIVAGHSVGEIAAYAIAGVIAADDAVALAATRGAEM
AKACATEPTGMSAVLGGDETEVLSRLEQLDLVPANRNAAGQIVAAGRLTALEKLAEDPPA
KARVRALGVAGAFHTEFMAPALDGFAAAAANIATADPTATLLSNRDGKPVTSAAAAMDTL
VSQLTQPVRWDLCTATLREHTVTAIVEFPPAGTLSGIAKRELRGVPARAVKSPADLDELA
NL [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: S275N |
[29] |
| Mutation Frequency |
4 out of 38 isolates |
|
| Target Name |
Bacterial Transcriptional regulator (FurA) |
Target Info |
| Gene Name |
furA |
| Uniprot ID |
FURA_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
SIPDYAEQLRTADLRVTRPRVAVLEAVNAHPHADTETIFGAVRFALPDVSRQAVYDVLHA
LTAAGLVRKIQPSGSVARYESRVGDNHHHIVCRSCGVIADVDCAVGEAPCLTASDHNGFL
LDEAEVIYWGLCPDCSISDTSRSHP [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: S5P |
[29] |
|
| Target Name |
Bacterial Beta-ketoacyl-acyl carrier protein reductase (mabA) |
Target Info |
| Gene Name |
mabA |
| Uniprot ID |
FABG_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTATATEGAKPPFVSRSVLVTGGNRGIGLAIAQRLAADGHKVAVTHRGSGAPKGLFGVEC
DVTDSDAVDRAFTAVEEHQGPVEVLVSNAGLSADAFLMRMTEEKFEKVINANLTGAFRVA
QRASRSMQRNKFGRMIFIGSVSGSWGIGNQANYAASKAGVIGMARSIARELSKANVTANV
VAPGYIDTDMTRALDERIQQGALQFIPAKRVGTPAEVAGVVSFLASEDASYISGAVIPVD
GGMGMGH [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: T21A |
[2] |
| Mutation Frequency |
3 out of 51 isolates in all mabA mutations |
|
| Mutation info |
Missense: A5P |
[46] |
|
| Mutation info |
Missense: V14L |
[46] |
|
| Target Name |
Bacterial Probable arabinosyltransferase C (embB) |
Target Info |
| Gene Name |
embB |
| Uniprot ID |
EMBB_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTQCASRRKSTPNRAILGAFASARGTRWVATIAGLIGFVLSVATPLLPVVQTTAMLDWPQ
RGQLGSVTAPLISLTPVDFTATVPCDVVRAMPPAGGVVLGTAPKQGKDANLQALFVVVSA
QRVDVTDRNVVILSVPREQVTSPQCQRIEVTSTHAGTFANFVGLKDPSGAPLRSGFPDPN
LRPQIVGVFTDLTGPAPPGLAVSATIDTRFSTRPTTLKLLAIIGAIVATVVALIALWRLD
QLDGRGSIAQLLLRPFRPASSPGGMRRLIPASWRTFTLTDAVVIFGFLLWHVIGANSSDD
GYILGMARVADHAGYMSNYFRWFGSPEDPFGWYYNLLALMTHVSDASLWMRLPDLAAGLV
CWLLLSREVLPRLGPAVEASKPAYWAAAMVLLTAWMPFNNGLRPEGIIALGSLVTYVLIE
RSMRYSRLTPAALAVVTAAFTLGVQPTGLIAVAALVAGGRPMLRILVRRHRLVGTLPLVS
PMLAAGTVILTVVFADQTLSTVLEATRVRAKIGPSQAWYTENLRYYYLILPTVDGSLSRR
FGFLITALCLFTAVFIMLRRKRIPSVARGPAWRLMGVIFGTMFFLMFTPTKWVHHFGLFA
AVGAAMAALTTVLVSPSVLRWSRNRMAFLAALFFLLALCWATTNGWWYVSSYGVPFNSAM
PKIDGITVSTIFFALFAIAAGYAAWLHFAPRGAGEGRLIRALTTAPVPIVAGFMAAVFVA
SMVAGIVRQYPTYSNGWSNVRAFVGGCGLADDVLVEPDTNAGFMKPLDGDSGSWGPLGPL
GGVNPVGFTPNGVPEHTVAEAIVMKPNQPGTDYDWDAPTKLTSPGINGSTVPLPYGLDPA
RVPLAGTYTTGAQQQSTLVSAWYLLPKPDDGHPLVVVTAAGKIAGNSVLHGYTPGQTVVL
EYAMPGPGALVPAGRMVPDDLYGEQPKAWRNLRFARAKMPADAVAVRVVAEDLSLTPEDW
IAVTPPRVPDLRSLQEYVGSTQPVLLDWAVGLAFPCQQPMLHANGIAEIPKFRITPDYSA
KKLDTDTWEDGTNGGLLGITDLLLRAHVMATYLSRDWARDWGSLRKFDTLVDAPPAQLEL
GTATRSGLWSPGKIRIGP [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: Y333H |
[45] |
|
| Target Name |
Bacterial LysR family transcriptional regulator (oxyR) |
Target Info |
| Gene Name |
oxyR |
| Species |
Mycobacterium tuberculosis |
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Frameshift: Plus 22 |
[50] |
|
| References |
| REF 1 |
Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996 Jan;173(1):196-202.
|
| REF 2 |
Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006 Aug;50(8):2640-9.
|
| REF 3 |
Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob Agents Chemother. 1999 Aug;43(8):2087-9.
|
| REF 4 |
Bioinformatics of antimicrobial resistance in the age of molecular epidemiology. Curr Opin Microbiol. 2015 Oct;27:45-50.
|
| REF 5 |
CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017 Jan 4;45(D1):D566-D573.
|
| REF 6 |
Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79(1):3-29.
|
| REF 7 |
Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J Clin Microbiol. 2005 Nov;43(11):5477-82.
|
| REF 8 |
Exclusive mutations related to isoniazid and ethionamide resistance among Mycobacterium tuberculosis isolates from Korea. Int J Tuberc Lung Dis. 2000 May;4(5):441-7.
|
| REF 9 |
Pyrazinamide-monoresistant Mycobacterium tuberculosis in the United States. J Clin Microbiol. 2001 Feb;39(2):647-50.
|
| REF 10 |
Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2001 Jul;45(7):2157-9.
|
| REF 11 |
Isoniazid activation defects in recombinant Mycobacterium tuberculosis catalase-peroxidase (KatG) mutants evident in InhA inhibitor production. Antimicrob Agents Chemother. 2003 Feb;47(2):670-5.
|
| REF 12 |
ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2003 Dec;47(12):3799-805.
|
| REF 13 |
Correlation between isoniazid resistance and superoxide reactivity in mycobacterium tuberculosis KatG. J Am Chem Soc. 2005 Sep 28;127(38):13428-42.
|
| REF 14 |
Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2006 Jun;2(6):e61.
|
| REF 15 |
Characterization of ndh gene of isoniazid resistant and susceptible Mycobacterium tuberculosis isolates from Brazil. Mem Inst Oswaldo Cruz. 2007 Feb;102(1):59-61.
|
| REF 16 |
Detection of mutation in isoniazid-resistant Mycobacterium tuberculosis isolates from tuberculosis patients in Belarus. Indian J Med Microbiol. 2008 Apr-Jun;26(2):143-7.
|
| REF 17 |
Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob Agents Chemother. 1997 Jul;41(7):1601-3.
|
| REF 18 |
Biological and molecular characteristics of Mycobacterium tuberculosis clinical isolates with low-level resistance to isoniazid in Japan. J Clin Microbiol. 2008 Jul;46(7):2263-8.
|
| REF 19 |
Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1995 Nov;39(11):2472-7.
|
| REF 20 |
katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob Agents Chemother. 1996 Sep;40(9):2187-9.
|
| REF 21 |
Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol Microbiol. 1996 Nov;22(3):583-92.
|
| REF 22 |
Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science. 1998 Jun 5;280(5369):1607-10.
|
| REF 23 |
Mechanisms of isoniazid resistance in Mycobacterium tuberculosis: enzymatic characterization of enoyl reductase mutants identified in isoniazid-resistant clinical isolates. J Infect Dis. 1998 Sep;178(3):769-75.
|
| REF 24 |
Rapid identification of a point mutation of the Mycobacterium tuberculosis catalase-peroxidase (katG) gene associated with isoniazid resistance. J Infect Dis. 1995 Jan;171(1):240-5.
|
| REF 25 |
Performance assessment of the GenoType MTBDRplus test and DNA sequencing in detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2009 Aug;47(8):2520-4.
|
| REF 26 |
Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995 Jan;15(2):235-45.
|
| REF 27 |
Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J Antimicrob Chemother. 2007 May;59(5):866-73.
|
| REF 28 |
Molecular characterization of drug-resistant Mycobacterium tuberculosis isolates from Ontario, Canada. J Antimicrob Chemother. 2009 Aug;64(2):263-6.
|
| REF 29 |
Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003 Apr;47(4):1241-50.
|
| REF 30 |
Molecular characterization of isoniazid resistance in Mycobacterium tuberculosis: identification of a novel mutation in inhA. Antimicrob Agents Chemother. 2006 Mar;50(3):1075-8.
|
| REF 31 |
New isoniazid/ethionamide resistance gene mutation and screening for multidrug-resistant Mycobacterium tuberculosis strains. Lancet. 1995 Aug 19;346(8973):502-3.
|
| REF 32 |
Molecular evidence for heterogeneity of the multiple-drug-resistant Mycobacterium tuberculosis population in Scotland (1990 to 1997). J Clin Microbiol. 1999 Apr;37(4):998-1003.
|
| REF 33 |
Genomic mutations in the katG, inhA and aphC genes are useful for the prediction of isoniazid resistance in Mycobacterium tuberculosis isolates from Kwazulu Natal, South Africa. Tuber Lung Dis. 2000;80(1):47-56.
|
| REF 34 |
Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob Agents Chemother. 2002 Feb;46(2):443-50.
|
| REF 35 |
Mutations in katG, inhA, and ahpC genes of Brazilian isoniazid-resistant isolates of Mycobacterium tuberculosis. J Clin Microbiol. 2003 Sep;41(9):4471-4.
|
| REF 36 |
Molecular analysis of isoniazid resistance in Mycobacterium tuberculosis isolates recovered from South Korea. Diagn Microbiol Infect Dis. 2003 Nov;47(3):497-502.
|
| REF 37 |
Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J Med Microbiol. 2004 Feb;53(Pt 2):107-13.
|
| REF 38 |
Three-dimensional model and molecular mechanism of Mycobacterium tuberculosis catalase-peroxidase (KatG) and isoniazid-resistant KatG mutants. Microb Drug Resist. 2004 Winter;10(4):269-79.
|
| REF 39 |
Molecular analysis of isoniazid-resistant Mycobacterium tuberculosis isolates from England and Wales reveals the phylogenetic significance of the ahpC -46A polymorphism. Antimicrob Agents Chemother. 2005 Apr;49(4):1455-64.
|
| REF 40 |
Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis isolates collected in Australia. Antimicrob Agents Chemother. 2005 Oct;49(10):4068-74.
|
| REF 41 |
Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med. 2006 Sep;12(9):1027-9.
|
| REF 42 |
Association of specific mutations in katG, rpoB, rpsL and rrs genes with spoligotypes of multidrug-resistant Mycobacterium tuberculosis isolates in... Clin Microbiol Infect. 2007 Jun;13(6):620-6.
|
| REF 43 |
Multiple mutations in katG and inhA identified in Thai isoniazid-resistant Mycobacterium tuberculosis isolates. Southeast Asian J Trop Med Public Health. 2007 Mar;38(2):376-82.
|
| REF 44 |
Development and evaluation of a line probe assay for rapid identification of pncA mutations in pyrazinamide-resistant mycobacterium tuberculosis st... J Clin Microbiol. 2007 Sep;45(9):2802-7.
|
| REF 45 |
Lack of correlation between embB mutation and ethambutol MIC in Mycobacterium tuberculosis clinical isolates from China. Antimicrob Agents Chemother. 2007 Dec;51(12):4515-7.
|
| REF 46 |
Molecular analysis of isoniazid resistance in different genotypes of Mycobacterium tuberculosis isolates from Iran. Microb Drug Resist. 2008 Dec;14(4):273-9.
|
| REF 47 |
Genotypic analysis of genes associated with isoniazid and ethionamide resistance in MDR-TB isolates from Thailand. Clin Microbiol Infect. 2010 Apr;16(4):396-9.
|
| REF 48 |
Mutations in rpoB and katG genes in Mycobacterium isolates from the Southeast of Mexico. Mem Inst Oswaldo Cruz. 2009 May;104(3):468-72.
|
| REF 49 |
Mutations in katG gene sequences in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis are rare. Antimicrob Agents Chemother. 1995 Oct;39(10):2276-81.
|
| REF 50 |
Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997 Mar;41(3):600-6.
|
| REF 51 |
A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob Agents Chemother. 1998 Sep;42(9):2443-5.
|


