| Drug General Information |
| Drug ID |
D0G3DL
|
| Drug Name |
Rifampin |
|
| Synonyms |
Abrifam; Archidyn; Arficin; Arzide; Benemicin; Benemycin; Dipicin; Doloresum; Eremfat; Famcin; Fenampicin; RFP; RMP; Ramp; Rifa; Rifadin; Rifadine; Rifagen; Rifaldazin; Rifaldazine; Rifaldin; Rifam; Rifamor; Rifampicin; Rifampicina; Rifampicine; Rifampicinum; Rifamsolin; Rifaprodin; Rifcin; Rifinah; Rifobac; Rifoldin; Rifoldine; Riforal; Rimactan; Rimactane; Rimactazid; Rimactizid; Rimazid; Rimycin; Sinerdol; Tubocin; Rifamicin AMP; Rifampicin SV; Rifampicine [French]; Rifampin [USAN]; Rifamycin AMP; Ba 41166; AZT + Rifampin; BA-41166E; Ba 41166/E; DRG-0109; Dione 21-acetate; L-5103; L-5103 Lepetit; Piperine & Rifampicin; R-Cin; R/AMP; Reserpine & Rifampicin; Rifadin (TN); Rifadin I.V; Rifampicin & EEP; Rifampicin & Propolis; Rifampicina [INN-Spanish]; Rifampicinum [INN-Latin]; Rifampin (USP); Rimactan (TN); Rimactane (TN); Rimycin (TN); Sinerdol (TN); Tubocin (TN); Rifadin I.V.; Rifampicin (JP15/INN); Rifampicin[INN:BAN:JAN]; Rifadin, Rimactane, Rifampicin, Rifampin; 1-b]furan-21-yl acetate; 3-(((4-Methyl-1-piperazinyl)imino)-methyl)rifamycin; 3-(((4-Methyl-1-piperazinyl)imino)methyl)rifamycin SV; 3-(4-Methylpiperazinyliminomethyl)-rifamycin SV; 3-(4-Methylpiperazinyliminomethyl)rifamycin SV; 3-([(4-Methyl-1-piperazinyl)imino]methyl)rifamycin SV; 3-[(4-Methyl-1-piperazinyl)iminomethyl]rifamycin SV; 3-[[(4-Methyl-1-piperazinyl)imino]-methyl]rifamycin; 8-(((4-Methyl-1-piperazinyl)imino)methyl)rifamycin SV; 8-(4-Methylpiperazinyliminomethyl) rifamycin SV; 8-[[(4-Methyl-1-piperazinyl)imino[methyl]rifamycin; 8-[[(4-Methyl-1-piperazinyl)imino]methyl]rifamycin sv; 8-[[(4-Methylpiperazinyl)imino]methyl]rifamycin sv; 8CI) |
| Drug Type |
Small molecular drug |
| Therapeutic Class |
Antituberculosis Agents |
| Company |
Norvatis Phamaceuticals Corporation |
| Structure |
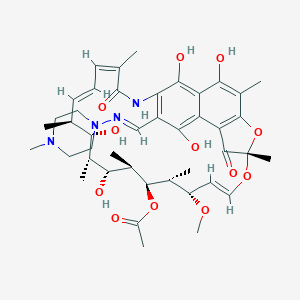
|
| Drug Resistance Mutations |
| Target Name |
Bacterial Probable arabinosyltransferase C (embB) |
Target Info |
| Gene Name |
embB |
| Uniprot ID |
EMBB_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MTQCASRRKSTPNRAILGAFASARGTRWVATIAGLIGFVLSVATPLLPVVQTTAMLDWPQ
RGQLGSVTAPLISLTPVDFTATVPCDVVRAMPPAGGVVLGTAPKQGKDANLQALFVVVSA
QRVDVTDRNVVILSVPREQVTSPQCQRIEVTSTHAGTFANFVGLKDPSGAPLRSGFPDPN
LRPQIVGVFTDLTGPAPPGLAVSATIDTRFSTRPTTLKLLAIIGAIVATVVALIALWRLD
QLDGRGSIAQLLLRPFRPASSPGGMRRLIPASWRTFTLTDAVVIFGFLLWHVIGANSSDD
GYILGMARVADHAGYMSNYFRWFGSPEDPFGWYYNLLALMTHVSDASLWMRLPDLAAGLV
CWLLLSREVLPRLGPAVEASKPAYWAAAMVLLTAWMPFNNGLRPEGIIALGSLVTYVLIE
RSMRYSRLTPAALAVVTAAFTLGVQPTGLIAVAALVAGGRPMLRILVRRHRLVGTLPLVS
PMLAAGTVILTVVFADQTLSTVLEATRVRAKIGPSQAWYTENLRYYYLILPTVDGSLSRR
FGFLITALCLFTAVFIMLRRKRIPSVARGPAWRLMGVIFGTMFFLMFTPTKWVHHFGLFA
AVGAAMAALTTVLVSPSVLRWSRNRMAFLAALFFLLALCWATTNGWWYVSSYGVPFNSAM
PKIDGITVSTIFFALFAIAAGYAAWLHFAPRGAGEGRLIRALTTAPVPIVAGFMAAVFVA
SMVAGIVRQYPTYSNGWSNVRAFVGGCGLADDVLVEPDTNAGFMKPLDGDSGSWGPLGPL
GGVNPVGFTPNGVPEHTVAEAIVMKPNQPGTDYDWDAPTKLTSPGINGSTVPLPYGLDPA
RVPLAGTYTTGAQQQSTLVSAWYLLPKPDDGHPLVVVTAAGKIAGNSVLHGYTPGQTVVL
EYAMPGPGALVPAGRMVPDDLYGEQPKAWRNLRFARAKMPADAVAVRVVAEDLSLTPEDW
IAVTPPRVPDLRSLQEYVGSTQPVLLDWAVGLAFPCQQPMLHANGIAEIPKFRITPDYSA
KKLDTDTWEDGTNGGLLGITDLLLRAHVMATYLSRDWARDWGSLRKFDTLVDAPPAQLEL
GTATRSGLWSPGKIRIGP [Mycobacterium tuberculosis]
|
| Targeted Disease |
Hansen disease; Staphylococcal infection; Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: Y319D |
[1], [2] |
|
| Mutation info |
Missense: Y333H |
[1], [2] |
|
| Target Name |
Bacterial DNA-directed RNA polymerase subunit beta (rpoB) |
Target Info |
| Gene Name |
rpoB |
| Uniprot ID |
RPOB_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MLEGCILADSRQSKTAASPSPSRPQSSSNNSVPGAPNRVSFAKLREPLEVPGLLDVQTDS
FEWLIGSPRWRESAAERGDVNPVGGLEEVLYELSPIEDFSGSMSLSFSDPRFDDVKAPVD
ECKDKDMTYAAPLFVTAEFINNNTGEIKSQTVFMGDFPMMTEKGTFIINGTERVVVSQLV
RSPGVYFDETIDKSTDKTLHSVKVIPSRGAWLEFDVDKRDTVGVRIDRKRRQPVTVLLKA
LGWTSEQIVERFGFSEIMRSTLEKDNTVGTDEALLDIYRKLRPGEPPTKESAQTLLENLF
FKEKRYDLARVGRYKVNKKLGLHVGEPITSSTLTEEDVVATIEYLVRLHEGQTTMTVPGG
VEVPVETDDIDHFGNRRLRTVGELIQNQIRVGMSRMERVVRERMTTQDVEAITPQTLINI
RPVVAAIKEFFGTSQLSQFMDQNNPLSGLTHKRRLSALGPGGLSRERAGLEVRDVHPSHY
GRMCPIETPEGPNIGLIGSLSVYARVNPFGFIETPYRKVVDGVVSDEIVYLTADEEDRHV
VAQANSPIDADGRFVEPRVLVRRKAGEVEYVPSSEVDYMDVSPRQMVSVATAMIPFLEHD
DANRALMGANMQRQAVPLVRSEAPLVGTGMELRAAIDAGDVVVAEESGVIEEVSADYITV
MHDNGTRRTYRMRKFARSNHGTCANQCPIVDAGDRVEAGQVIADGPCTDDGEMALGKNLL
VAIMPWEGHNYEDAIILSNRLVEEDVLTSIHIEEHEIDARDTKLGAEEITRDIPNISDEV
LADLDERGIVRIGAEVRDGDILVGKVTPKGETELTPEERLLRAIFGEKAREVRDTSLKVP
HGESGKVIGIRVFSREDEDELPAGVNELVRVYVAQKRKISDGDKLAGRHGNKGVIGKILP
VEDMPFLADGTPVDIILNTHGVPRRMNIGQILETHLGWCAHSGWKVDAAKGVPDWAARLP
DELLEAQPNAIVSTPVFDGAQEAELQGLLSCTLPNRDGDVLVDADGKAMLFDGRSGEPFP
YPVTVGYMYIMKLHHLVDDKIHARSTGPYSMITQQPLGGKAQFGGQRFGEMECWAMQAYG
AAYTLQELLTIKSDDTVGRVKVYEAIVKGENIPEPGIPESFKVLLKELQSLCLNVEVLSS
DGAAIELREGEDEDLERAAANLGINLSRNESASVEDLA [Mycobacterium tuberc
ulosis]
|
| Targeted Disease |
Hansen disease; Staphylococcal infection; Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Missense: H526L |
[3] |
| Mutation Frequency |
2% of the patients |
|
| Mutation info |
Missense: L533P |
[4], [5], [6] |
| Mutation Frequency |
15 out of 258 isolates |
|
| Mutation info |
Missense: L533P |
[4], [5], [6] |
| Mutation Frequency |
15 out of 258 isolates |
|
| Mutation info |
Missense: D516V |
[3], [6], [7] |
| Mutation Frequency |
1% of the patients |
|
| Mutation info |
Missense: M515I |
[6], [7], [8] |
|
| Mutation info |
Missense: F505L |
[6], [9], [10] |
| Mutation Frequency |
2 out of 41 isolates |
|
| Mutation info |
Missense: R528P |
[6], [11], [12] |
|
| Mutation info |
Missense: G507D |
[6], [13], [14] |
| Mutation Frequency |
2 out of 46 isolates |
|
| Mutation info |
Missense: H526Q |
[3], [6], [15] |
| Mutation Frequency |
1% of the isolates |
|
| Mutation info |
Missense: G523W |
[16], [17] |
| Mutation Frequency |
37 out of 42 isolates |
|
| Mutation info |
Missense: D516N |
[18], [6] |
|
| Mutation info |
Missense: E504A |
[18], [6] |
|
| Mutation info |
Missense: I572F |
[19], [6] |
|
| Mutation info |
Missense: S522Q |
[19], [6] |
|
| Mutation info |
Missense: H526G |
[8], [6] |
|
| Mutation info |
Missense: H526T |
[8], [6] |
|
| Mutation info |
Missense: L521M |
[8], [6] |
|
| Mutation info |
Missense: S512R |
[8], [6] |
|
| Mutation info |
Missense: S531C |
[8], [6] |
|
| Mutation info |
Missense: V146F |
[20], [6] |
|
| Mutation info |
Missense: V146F |
[20], [6] |
|
| Mutation info |
Insertion: F514 |
[4], [3] |
|
| Mutation info |
Deletion: F514 |
[4], [3] |
|
| Mutation info |
Missense: E672D |
[21], [6] |
|
| Mutation info |
Missense: L571V |
[21], [6] |
|
| Mutation info |
Missense: K527N |
[11], [6] |
|
| Mutation info |
Missense: L511V |
[11], [6] |
|
| Mutation info |
Missense: N518T |
[11], [6] |
|
| Mutation info |
Missense: R528H |
[11], [6] |
|
| Mutation info |
Missense: Q490H |
[22], [6] |
|
| Mutation info |
Missense: H526N |
[23], [6] |
|
| Mutation info |
Missense: Q517L |
[7], [6] |
|
| Mutation info |
Missense: S512I |
[7], [6] |
|
| Mutation info |
Missense: N518I |
[24], [6] |
|
| Mutation info |
Missense: S531F |
[24], [6] |
|
| Mutation info |
Missense: S531F |
[24], [6] |
|
| Mutation info |
Missense: Q513R |
[25], [6] |
| Mutation Frequency |
17 out of 840 isolates |
|
| Mutation info |
Missense: S522W |
[25], [6] |
|
| Mutation info |
Missense: D516G |
[26], [6] |
|
| Mutation info |
Missense: S509R |
[27], [6] |
|
| Mutation info |
Missense: F514L |
[28], [29] |
| Mutation Frequency |
1 out of 39 isolates |
|
| Mutation info |
Missense: L538P |
[30], [6] |
| Mutation Frequency |
101 out of 112 isolates in all rpoB mutations |
|
| Mutation info |
Missense: E541G |
[16], [31] |
|
| Mutation info |
Missense: H526S |
[31], [6] |
| Mutation Frequency |
49 out of 54 isolates |
|
| Mutation info |
Missense: Q513E |
[31], [6] |
| Mutation Frequency |
49 out of 54 isolates |
|
| Mutation info |
Missense: Q517H |
[32], [6] |
| Mutation Frequency |
97 out of 148 isolates |
|
| Mutation info |
Missense: P564L |
[33], [6] |
|
| Mutation info |
Missense: P564L |
[33], [6] |
|
| Mutation info |
Missense: S509Q |
[12], [6] |
|
| Mutation info |
Missense: G507S |
[13], [6] |
|
| Mutation info |
Missense: H526F |
[13], [6] |
|
| Mutation info |
Missense: N519K |
[13], [6] |
| Mutation Frequency |
2 out of 46 isolates |
|
| Mutation info |
Missense: H526P |
[15], [6] |
|
| Mutation info |
Missense: S522L |
[15], [6] |
|
| Mutation info |
Missense: S531W |
[15], [6] |
|
| Mutation info |
Missense: K527Q |
[34], [6] |
| Mutation Frequency |
1 out of 110 isolates |
|
| Mutation info |
Missense: S531L |
[35], [6] |
| Mutation Frequency |
1 out of 7 isolates |
|
| Mutation info |
Missense: H526D |
[3], [6] |
| Mutation Frequency |
52 out of 128 isolates |
|
| Mutation info |
Missense: L511P |
[3], [6] |
| Mutation Frequency |
3% of the isolates |
|
| Mutation info |
Missense: L511R |
[3], [6] |
| Mutation Frequency |
2% of the patients |
|
| Mutation info |
Missense: Q513K |
[3], [6] |
| Mutation Frequency |
1% of the isolates |
|
| Mutation info |
Deletion: Q517-N518 |
[24], [3] |
|
| Mutation info |
Missense: S512T |
[3], [6] |
| Mutation Frequency |
1% of the isolates |
|
| Mutation info |
Missense: H526Y |
[3], [6] |
| Mutation Frequency |
32% of the isolates |
|
| Mutation info |
Missense: H526Y |
[3], [6] |
| Mutation Frequency |
32% of the isolates |
|
| Mutation info |
Missense: Q513L |
[3], [6] |
| Mutation Frequency |
2% of the patients |
|
| Mutation info |
Missense: Q513L |
[3], [6] |
| Mutation Frequency |
2% of the patients |
|
| Mutation info |
Missense: A381V |
[29], [6] |
| Mutation Frequency |
1 out of 39 isolates |
|
| Mutation info |
Missense: L521P |
[29], [6] |
| Mutation Frequency |
1 out of 39 isolates |
|
| Mutation info |
Missense: H526C |
[14], [6] |
| Mutation Frequency |
2 out of 37 isolates |
|
| Mutation info |
Missense: H526R |
[14], [6] |
| Mutation Frequency |
1 out of 37 isolates |
|
| Mutation info |
Missense: N518H |
[14], [6] |
| Mutation Frequency |
1 out of 37 isolates |
|
| Mutation info |
Missense: Q513P |
[14], [6] |
| Mutation Frequency |
2 out of 37 isolates |
|
| Mutation info |
Missense: Q513P |
[14], [6] |
| Mutation Frequency |
2 out of 37 isolates |
|
| Mutation info |
Missense: T508S |
[36], [37] |
|
| Mutation info |
Missense: D516H |
[13], [6] |
| Mutation Frequency |
3 out of 46 isolates |
|
| Mutation info |
Missense: D516Y |
[3], [6] |
| Mutation Frequency |
2% of the patients |
|
| Mutation info |
Missense: S574L |
[38], [6] |
| Mutation Frequency |
25 out of 38 isolates in all rpoB mutations |
|
| Mutation info |
Missense: S553A |
[16] |
|
| Mutation info |
Missense: T525I |
[16] |
|
| Mutation info |
Missense: D516A |
[8] |
|
| Mutation info |
Deletion: G507-T508 |
[8] |
|
| Mutation info |
Deletion: Q513-F514 |
[8] |
|
| Mutation info |
Missense: Q513-M516 > H |
[8] |
|
| Mutation info |
Missense: Q517-N519 > H |
[8] |
|
| Mutation info |
Missense: S531Q |
[8] |
|
| Mutation info |
Deletion: F514-D516 |
[4] |
|
| Mutation info |
Missense: F514V |
[4] |
|
| Mutation info |
Deletion: K527 |
[4] |
|
| Mutation info |
Missense: L524W |
[4] |
|
| Mutation info |
Deletion: M515-N518 |
[4] |
|
| Mutation info |
Missense: Q513H |
[4] |
|
| Mutation info |
Missense: T525P |
[4] |
|
| Mutation info |
Missense: I561T |
[21] |
|
| Mutation info |
Missense: R633C |
[21] |
|
| Mutation info |
Deletion: Q517 |
[36] |
|
| Mutation info |
Missense: P535S |
[39] |
| Mutation Frequency |
4 out of 34 isolates |
|
| Mutation info |
Insertion: R515 |
[22] |
|
| Mutation info |
Missense: I561V |
[23] |
|
| Mutation info |
Missense: Q510H |
[7] |
|
| Mutation info |
Missense: L521L |
[25] |
| Mutation Frequency |
2 out of 840 isolates |
|
| Mutation info |
Missense: D516F |
[26] |
|
| Mutation info |
Insertion: F515 |
[26] |
|
| Mutation info |
Missense: N518D |
[38] |
| Mutation Frequency |
25 out of 38 isolates in all rpoB mutations |
|
| Mutation info |
Missense: S512G |
[38] |
| Mutation Frequency |
25 out of 38 isolates in all rpoB mutations |
|
| Mutation info |
Missense: F505S |
[30] |
|
| Mutation info |
Nonsense: L538 STOP |
[30] |
| Mutation Frequency |
101 out of 112 isolates in all rpoB mutations |
|
| Mutation info |
Missense: N518Y |
[30] |
| Mutation Frequency |
101 out of 112 isolates in all rpoB mutations |
|
| Mutation info |
Missense: P535H |
[31] |
|
| Mutation info |
Missense: S622A |
[31] |
|
| Mutation info |
Missense: L545M |
[40] |
| Mutation Frequency |
4 out of 14 isolates |
|
| Mutation info |
Missense: E562G |
[33] |
|
| Mutation info |
Missense: L524S |
[12] |
|
| Mutation info |
Missense: Q510K |
[12] |
| Mutation Frequency |
59 out of 409 isolates |
|
| Mutation info |
Missense: G513N |
[13] |
|
| Mutation info |
Missense: G523A |
[13] |
|
| Mutation info |
Nonsense: Q513 STOP |
[13] |
|
| Mutation info |
Missense: T508A |
[13] |
| Mutation Frequency |
35 out of 46 isolates |
|
| Mutation info |
Missense: T508H |
[13] |
| Mutation Frequency |
35 out of 46 isolates |
|
| Mutation info |
Missense: T508P |
[13] |
| Mutation Frequency |
35 out of 46 isolates |
|
| Mutation info |
Missense: S450L |
[41] |
|
| Mutation info |
Missense: S531Y |
[15] |
|
| Mutation info |
Missense: S509T |
[34] |
| Mutation Frequency |
1 out of 110 isolates |
|
| Mutation info |
Deletion: N519 |
[10] |
| Mutation Frequency |
2 out of 41 isolates |
|
| Mutation info |
Missense: D516E |
[3] |
| Mutation Frequency |
2% of the patients |
|
| Mutation info |
Deletion: D516-Q517 |
[3] |
|
| Mutation info |
Insertion: FM513 |
[3] |
|
| Mutation info |
Missense: A473T |
[42] |
|
| Mutation info |
Missense: A477T |
[42] |
|
| Mutation info |
Missense: H481N |
[42] |
|
| Mutation info |
Missense: L466S |
[42] |
|
| Mutation info |
Missense: Q465R |
[42] |
|
| Mutation info |
Missense: Q468K |
[42] |
|
| Mutation info |
Missense: D516K |
[6] |
|
| Mutation info |
Missense: D516T |
[6] |
|
| Mutation info |
Missense: H526E |
[6] |
|
| Mutation info |
Missense: L511M |
[6] |
|
| Mutation info |
Missense: L533R |
[6] |
|
| Mutation info |
Missense: L538R |
[6] |
|
| Mutation info |
Missense: M515V |
[6] |
|
| Mutation info |
Missense: Q513D |
[6] |
|
| Mutation info |
Missense: R529C |
[6] |
|
| Mutation info |
Missense: R529Q |
[6] |
|
| Mutation info |
Missense: R529S |
[6] |
|
| Mutation info |
Missense: R687H |
[6] |
|
| Mutation info |
Missense: S456L |
[6] |
|
| Mutation info |
Missense: S531G |
[6] |
|
| Mutation info |
Missense: T563P |
[6] |
|
| Mutation info |
Missense: V472I |
[6] |
|
| Mutation info |
Missense: R484H + E517Q |
[43] |
|
| References |
| REF 1 |
Lack of correlation between embB mutation and ethambutol MIC in Mycobacterium tuberculosis clinical isolates from China. Antimicrob Agents Chemother. 2007 Dec;51(12):4515-7.
|
| REF 2 |
Bioinformatics of antimicrobial resistance in the age of molecular epidemiology. Curr Opin Microbiol. 2015 Oct;27:45-50.
|
| REF 3 |
Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994 Apr;32(4):1095-8.
|
| REF 4 |
Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. J Clin Microbiol. 2000 Aug;38(8):3119-22.
|
| REF 5 |
Comparative antimycobacterial activities of rifampin, rifapentine, and KRM-1648 against a collection of rifampin-resistant Mycobacterium tuberculosis isolates with known rpoB mutations. Antimicrob Agents Chemother. 1996 Nov;40(11):2655-7.
|
| REF 6 |
CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017 Jan 4;45(D1):D566-D573.
|
| REF 7 |
Molecular characterization of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Poland. J Clin Microbiol. 2004 Jun;42(6):2425-31.
|
| REF 8 |
Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79(1):3-29.
|
| REF 9 |
In-depth molecular characterization of Mycobacterium tuberculosis from New Delhi--predominance of drug resistant isolates of the 'modern' (TbD1) type. PLoS One. 2009;4(2):e4540.
|
| REF 10 |
Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994 Jul 30;344(8918):293-8.
|
| REF 11 |
Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob Agents Chemother. 2002 Feb;46(2):443-50.
|
| REF 12 |
Mutations in rpoB and katG genes in Mycobacterium isolates from the Southeast of Mexico. Mem Inst Oswaldo Cruz. 2009 May;104(3):468-72.
|
| REF 13 |
High-level rifampin resistance correlates with multiple mutations in the rpoB gene of pulmonary tuberculosis isolates from the Afghanistan border of Iran. J Clin Microbiol. 2009 Sep;47(9):2744-50.
|
| REF 14 |
Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR-single-strand conformation polymorphism analysis for rifampin susceptibility testing. J Clin Microbiol. 1997 Feb;35(2):492-4.
|
| REF 15 |
Mutation position and type of substitution in the beta-subunit of the RNA polymerase influence in-vitro activity of rifamycins in rifampicin-resistant Mycobacterium tuberculosis. J Antimicrob Chemother. 1995 Feb;35(2):345-8.
|
| REF 16 |
rpoB mutations in multidrug-resistant strains of Mycobacterium tuberculosis isolated in Italy. J Clin Microbiol. 1999 Apr;37(4):1197-9.
|
| REF 17 |
Chronic cases of tuberculosis in Casablanca, Morocco. Int J Tuberc Lung Dis. 2003 Jul;7(7):660-4.
|
| REF 18 |
Universal pattern of RpoB gene mutations among multidrug-resistant isolates of Mycobacterium tuberculosis complex from Africa. Int J Tuberc Lung Dis. 1999 Jul;3(7):620-6.
|
| REF 19 |
Bacteriological and molecular analysis of rifampin-resistant Mycobacterium tuberculosis strains isolated in Australia. J Clin Microbiol. 1999 Dec;37(12):3844-50.
|
| REF 20 |
Mutations in the beginning of the rpoB gene can induce resistance to rifamycins in both Helicobacter pylori and Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2000 Apr;44(4):1075-7.
|
| REF 21 |
Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2001 Jan;39(1):107-10.
|
| REF 22 |
Characterization of rpoB mutations in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Turkey by DNA sequencing and line probe assay. J Clin Microbiol. 2002 Dec;40(12):4435-8.
|
| REF 23 |
Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J Med Microbiol. 2004 Feb;53(Pt 2):107-13.
|
| REF 24 |
Disequilibrium in distribution of resistance mutations among Mycobacterium tuberculosis Beijing and non-Beijing strains isolated from patients in Germany. Antimicrob Agents Chemother. 2005 Mar;49(3):1229-31.
|
| REF 25 |
Analysis of rpoB and pncA mutations in the published literature: an insight into the role of oxidative stress in Mycobacterium tuberculosis evolution J Antimicrob Chemother. 2005 May;55(5):674-9.
|
| REF 26 |
Rifampicin and isoniazid resistance mutations in Mycobacterium tuberculosis strains isolated from patients in Kazakhstan. Int J Tuberc Lung Dis. 2005 Oct;9(10):1161-7.
|
| REF 27 |
Detection of multidrug resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2007 Jan;45(1):179-92.
|
| REF 28 |
Association of specific mutations in katG, rpoB, rpsL and rrs genes with spoligotypes of multidrug-resistant Mycobacterium tuberculosis isolates in... Clin Microbiol Infect. 2007 Jun;13(6):620-6.
|
| REF 29 |
Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol Lett. 1996 Oct 15;144(1):103-8.
|
| REF 30 |
A newly identified 191A/C mutation in the Rv2629 gene that was significantly associated with rifampin resistance in Mycobacterium tuberculosis. J Proteome Res. 2007 Dec;6(12):4564-71.
|
| REF 31 |
Distribution of rpoB mutations among multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin Microbiol Infect. 2008 May;14(5):446-53.
|
| REF 32 |
Identification of rifampin-resistant genotypes in Mycobacterium tuberculosis by PCR-reverse dot blot hybridization. Mol Biotechnol. 2009 Jan;41(1):1-7.
|
| REF 33 |
Comparison of two commercial assays for the characterization of rpoB mutations in Mycobacterium tuberculosis and description of new mutations conferring weak resistance to rifampicin. J Antimicrob Chemother. 2009 Aug;64(2):259-62.
|
| REF 34 |
Characterization of rifampin-resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994 Oct;38(10):2380-6.
|
| REF 35 |
Isolation of the gene for the beta subunit of RNA polymerase from rifampicin-resistant Mycobacterium tuberculosis and identification of new mutations. Am J Respir Cell Mol Biol. 1994 Dec;11(6):639-43.
|
| REF 36 |
Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J Clin Microbiol. 2001 Aug;39(8):2987-90.
|
| REF 37 |
Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J Clin Microbiol. 1998 Jan;36(1):20-3.
|
| REF 38 |
Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J Antimicrob Chemother. 2007 May;59(5):866-73.
|
| REF 39 |
Mutations in the rpoB and katG genes leading to drug resistance in Mycobacterium tuberculosis in Latvia. J Clin Microbiol. 2002 Oct;40(10):3789-92.
|
| REF 40 |
Genotypic analysis of isoniazid and rifampin resistance in drug-resistant clinical Mycobacterium tuberculosis complex isolates in southern Turkey. Jpn J Infect Dis. 2008 Jul;61(4):255-60.
|
| REF 41 |
Longitudinal whole genome analysis of pre and post drug treatment Mycobacterium tuberculosis isolates reveals progressive steps to drug resistance. Tuberculosis (Edinb). 2016 May;98:50-5.
|
| REF 42 |
The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013 Jul;57(7):3348-57.
|
| REF 43 |
Mutations in rpoB and fusA cause resistance to rifampicin and fusidic acid in methicillin-resistant Staphylococcus aureus strains from a tertiary hospital in Malaysia.Trans R Soc Trop Med Hyg.2014 Feb;108(2):112-8.
|


