Drug Information
| Drug General Information | Top | |||
|---|---|---|---|---|
| Drug ID |
D0N6RF
|
|||
| Former ID |
DAP000766
|
|||
| Drug Name |
Telmisartan
|
|||
| Synonyms |
Kinzal; Kinzalmono; Micardis; Pritor; Abbott brand of telmisartan; Boehringer Ingelheim brand of telmisartan; Glaxo Wellcome brand of telmisartan; GlaxoSmithKline brand of telmisartan; BIBR 277; BIBR 277SE; BIBR-277; BIBR-277SE; Bay 68-9291; Micardis (TN); Telmisartan [USAN:INN]; YM-086; BIBR-277-SE; Telmisartan (JAN/USAN/INN); Micardis, Targit, Temax, BIBR277, Telmisartan; 2-[4-[[4-methyl-6-(1-methylbenzimidazol-2-yl)-2-propylbenzimidazol-1-yl]methyl]phenyl]benzoic acid; 4'-((1,4'-dimethyl-2'-propyl(2,6'-bi-1H-benzimidazol)-1'-yl)methyl)-(1,1'-biphenyl)-2-carboxylic acid; 4'-((4-Methyl-6-(1-methyl-2-benzimidazolyl)-2-propyl-1-benzimidazolyl)methyl)-2-biphenylcarboxylic acid; 4'-((4-mehtyl-6-(1-methyl-2-benzimidazolyl)-2-propyl-1-benzimmidazolyl)methyl)-2-biphenylcarboxylic acid; 4'-[(1,4'-dimethyl-2'propyl[2,6'-bi-1H-benzimidazol]-1'-yl)methyl]-[1,1'-biphenyl]-2-carboxylic acid; 4'-[(1,7'-dimethyl-2'-propyl-1H,3'H-2,5'-bibenzimidazol-3'-yl)methyl][1,1'-biphenyl]-2-carboxylic acid; 4'-[(1,7'-dimethyl-2'-propyl-1H,3'H-2,5'-bibenzimidazol-3'-yl)methyl]biphenyl-2-carboxylic acid
Click to Show/Hide
|
|||
| Drug Type |
Small molecular drug
|
|||
| Indication | Hypertension [ICD-11: BA00-BA04; ICD-9: 401] | Approved | [1], [2] | |
| Therapeutic Class |
Antihypertensive Agents
|
|||
| Company |
Boehringer Ingelheim
|
|||
| Structure |
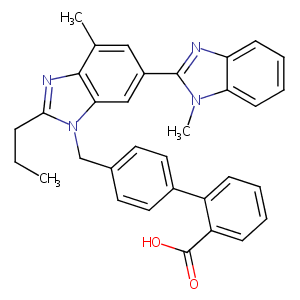 |
Download2D MOL |
||
| Formula |
C33H30N4O2
|
|||
| Canonical SMILES |
CCCC1=NC2=C(N1CC3=CC=C(C=C3)C4=CC=CC=C4C(=O)O)C=C(C=C2C)C5=NC6=CC=CC=C6N5C
|
|||
| InChI |
1S/C33H30N4O2/c1-4-9-30-35-31-21(2)18-24(32-34-27-12-7-8-13-28(27)36(32)3)19-29(31)37(30)20-22-14-16-23(17-15-22)25-10-5-6-11-26(25)33(38)39/h5-8,10-19H,4,9,20H2,1-3H3,(H,38,39)
|
|||
| InChIKey |
RMMXLENWKUUMAY-UHFFFAOYSA-N
|
|||
| CAS Number |
CAS 144701-48-4
|
|||
| PubChem Compound ID | ||||
| PubChem Substance ID |
9912, 6866718, 7847693, 7980737, 8190092, 11372057, 11374980, 11485867, 11489792, 11490904, 11493075, 11528632, 12014783, 14860450, 26612858, 26680963, 26719833, 26749005, 26749006, 43122721, 46386620, 46505370, 46530547, 47350860, 47796474, 48416595, 49648460, 49830991, 50107502, 56312042, 56313977, 57315885, 79018553, 81041072, 81092854, 85209733, 85789678, 87225392, 92124871, 92308092, 92308546, 92309313, 92712346, 93166523, 103271997, 103974749, 104253350, 104335216, 117546129, 117867825
|
|||
| ChEBI ID |
CHEBI:9434
|
|||
| ADReCS Drug ID | BADD_D02147 | |||
| SuperDrug ATC ID |
C09CA07
|
|||
| SuperDrug CAS ID |
cas=144701484
|
|||
| Interaction between the Drug and Microbe | Top | |||
|---|---|---|---|---|
| The Metabolism of Drug Affected by Studied Microbe(s) | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Bacteroides cellulosilyticus DSM 14838
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides cellulosilyticus DSM 14838 (log2FC = -0.567; p = 0.035). | |||
|
Studied Microbe: Bacteroides dorei DSM 17855
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides dorei DSM 17855 (log2FC = -0.999; p = 0.006). | |||
|
Studied Microbe: Bacteroides fragilis HMW 610
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides fragilis HMW 610 (log2FC = -0.615; p = 0.017). | |||
|
Studied Microbe: Bacteroides fragilis NCTC 9343
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides fragilis NCTC 9343 (log2FC = -0.886; p = 0.041). | |||
|
Studied Microbe: Bacteroides ovatus ATCC 8483
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides ovatus ATCC 8483 (log2FC = -0.349; p = 0.029). | |||
|
Studied Microbe: Bacteroides stercoris ATCC 43183
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides stercoris ATCC 43183 (log2FC = -0.599; p = 0.04). | |||
|
Studied Microbe: Bacteroides thetaiotaomicron 7330
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides thetaiotaomicron 7330 (log2FC = -0.619; p = 0.022). | |||
|
Studied Microbe: Bacteroides uniformis ATCC 8492
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides uniformis ATCC 8492 (log2FC = -1.005; p = 0.009). | |||
|
Studied Microbe: Bacteroides vulgatus ATCC 8482
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides vulgatus ATCC 8482 (log2FC = -0.856; p = 0.026). | |||
|
Studied Microbe: Bacteroides xylanisolvens DSM18836
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Bacteroides xylanisolvens DSM18836 (log2FC = -0.784; p = 0.03). | |||
|
Studied Microbe: Odoribacter splanchnicus
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Odoribacter splanchnicus (log2FC = -0.402; p = 0.035). | |||
|
Studied Microbe: Parabacteroides distasonis ATCC 8503
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Telmisartan can be metabolized by Parabacteroides distasonis ATCC 8503 (log2FC = -0.73; p = 0.034). | |||
| The Abundace of Studied Microbe(s) Regulated by Drug | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Parabacteroides distasonis
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Parabacteroides distasonis was decreased by Telmisartan (adjusted p-values: 4.47E-03). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Eubacteriales | ||||
|
Studied Microbe: Clostridioides difficile
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Clostridioides difficile was decreased by Telmisartan (adjusted p-values: 1.63E-05). | |||
|
Studied Microbe: Clostridium perfringens
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Clostridium perfringens was decreased by Telmisartan (adjusted p-values: 7.24E-03). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Lactobacillales | ||||
|
Studied Microbe: Streptococcus salivarius
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Streptococcus salivarius was decreased by Telmisartan (adjusted p-values: 6.93E-03). | |||
| Target and Pathway | Top | |||
|---|---|---|---|---|
| Target(s) | Angiotensin II receptor type-1 (AGTR1) | Target Info | Antagonist | [5] |
| KEGG Pathway | Calcium signaling pathway | |||
| cGMP-PKG signaling pathway | ||||
| Neuroactive ligand-receptor interaction | ||||
| Adrenergic signaling in cardiomyocytes | ||||
| Vascular smooth muscle contraction | ||||
| Renin-angiotensin system | ||||
| Renin secretion | ||||
| Pathways in cancer | ||||
| NetPath Pathway | TGF_beta_Receptor Signaling Pathway | |||
| Panther Pathway | Angiotensin II-stimulated signaling through G proteins and beta-arrestin | |||
| Pathwhiz Pathway | Angiotensin Metabolism | |||
| Muscle/Heart Contraction | ||||
| Pathway Interaction Database | Arf6 trafficking events | |||
| Arf6 signaling events | ||||
| Angiopoietin receptor Tie2-mediated signaling | ||||
| Reactome | Peptide ligand-binding receptors | |||
| G alpha (q) signalling events | ||||
| WikiPathways | ACE Inhibitor Pathway | |||
| GPCRs, Class A Rhodopsin-like | ||||
| Gastrin-CREB signalling pathway via PKC and MAPK | ||||
| Peptide GPCRs | ||||
| Allograft Rejection | ||||
| GPCR ligand binding | ||||
| GPCR downstream signaling | ||||
| References | Top | |||
|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 592). | |||
| REF 2 | Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007 Mar;70(3):461-77. | |||
| REF 3 | Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019 Jun;570(7762):462-467. | |||
| REF 4 | Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018 Mar 29;555(7698):623-628. | |||
| REF 5 | Deletion of angiotensin II type I receptor reduces hepatic steatosis. J Hepatol. 2009 Jun;50(6):1226-35. | |||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

