Drug Information
| Drug General Information | Top | |||
|---|---|---|---|---|
| Drug ID |
D0Z4EI
|
|||
| Former ID |
DIB010980
|
|||
| Drug Name |
Mifepristone
|
|||
| Synonyms |
Mifedren; HRA-052015; Mifepristone (Cushing's disease); ACTH modulator (Cushing's disease), HRA; Adrenocorticotropic hormone modulator (Cushing's disease), HRA Pharma; Mifepristone (Cushing's disease), HRA pharma
Click to Show/Hide
|
|||
| Drug Type |
Small molecular drug
|
|||
| Indication | Cushing disease [ICD-11: 5A70; ICD-10: E24; ICD-9: 255] | Approved | [1], [2], [3] | |
| Company |
HRA Pharma
|
|||
| Structure |
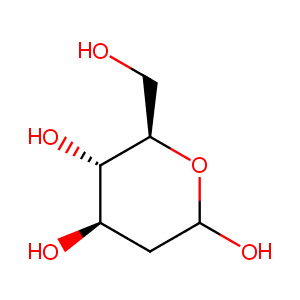 |
Download2D MOL |
||
| Formula |
C29H35NO2
|
|||
| Canonical SMILES |
CC#CC1(CCC2C1(CC(C3=C4CCC(=O)C=C4CCC23)C5=CC=C(C=C5)N(C)C)C)O
|
|||
| InChI |
1S/C29H35NO2/c1-5-15-29(32)16-14-26-24-12-8-20-17-22(31)11-13-23(20)27(24)25(18-28(26,29)2)19-6-9-21(10-7-19)30(3)4/h6-7,9-10,17,24-26,32H,8,11-14,16,18H2,1-4H3/t24-,25+,26-,28-,29-/m0/s1
|
|||
| InChIKey |
VKHAHZOOUSRJNA-GCNJZUOMSA-N
|
|||
| CAS Number |
CAS 84371-65-3
|
|||
| PubChem Compound ID | ||||
| PubChem Substance ID |
9854, 610895, 827617, 7847651, 7885329, 7979988, 8184037, 10321543, 11466327, 11467447, 11486193, 11533034, 12013919, 12146248, 14758749, 14905326, 17405362, 24278572, 26752199, 26759172, 34719152, 46386955, 46505795, 46518693, 47573358, 47720589, 47795017, 47795018, 48018884, 48318385, 48393908, 48416281, 49698423, 49835339, 49965772, 50124354, 50576278, 53777907, 53788556, 56312231, 56313355, 56313657, 56313765, 56352854, 56424093, 57313727, 85752924, 85789352, 90341299, 92125204
|
|||
| ChEBI ID |
CHEBI:50692
|
|||
| ADReCS Drug ID | BADD_D01463 | |||
| Interaction between the Drug and Microbe | Top | |||
|---|---|---|---|---|
| The Metabolism of Drug Affected by Studied Microbe(s) | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Bacteroides dorei DSM 17855
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Bacteroides dorei DSM 17855 (log2FC = -0.778; p = 0.005). | |||
|
Studied Microbe: Bacteroides fragilis ATCC43859
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Bacteroides fragilis ATCC43859 (log2FC = -0.458; p = 0.041). | |||
|
Studied Microbe: Bacteroides fragilis HMW 610
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Bacteroides fragilis HMW 610 (log2FC = -0.554; p = 0.01). | |||
|
Studied Microbe: Bacteroides fragilis NCTC 9343
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Bacteroides fragilis NCTC 9343 (log2FC = -0.504; p = 0.045). | |||
|
Studied Microbe: Bacteroides fragilis str. DS-208
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Bacteroides fragilis str. DS-208 (log2FC = -0.355; p = 0.018). | |||
|
Studied Microbe: Bacteroides vulgatus ATCC 8482
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Bacteroides vulgatus ATCC 8482 (log2FC = -0.713; p = 0.001). | |||
|
Studied Microbe: Odoribacter splanchnicus
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Odoribacter splanchnicus (log2FC = -0.348; p = 0.012). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Eubacteriales | ||||
|
Studied Microbe: Blautia hansenii DSM20583
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Blautia hansenii DSM20583 (log2FC = -0.492; p = 0.006). | |||
|
Studied Microbe: Clostridium sp.
Show/Hide Hierarchy
|
[4] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Mifepristone can be metabolized by Clostridium sp. (log2FC = -0.476; p = 0.006). | |||
| The Abundace of Studied Microbe(s) Regulated by Drug | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Bacteroides uniformis
Show/Hide Hierarchy
|
[5] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Bacteroides uniformis was decreased by Mifepristone (adjusted p-values: 2.82E-03). | |||
|
Studied Microbe: Parabacteroides merdae
Show/Hide Hierarchy
|
[5] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Parabacteroides merdae was decreased by Mifepristone (adjusted p-values: 5.97E-05). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Eubacteriales | ||||
|
Studied Microbe: Clostridioides difficile
Show/Hide Hierarchy
|
[5] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Clostridioides difficile was decreased by Mifepristone (adjusted p-values: 4.04E-04). | |||
| Target and Pathway | Top | |||
|---|---|---|---|---|
| Target(s) | Glucocorticoid receptor (NR3C1) | Target Info | Modulator | [6] |
| KEGG Pathway | Neuroactive ligand-receptor interaction | |||
| NetPath Pathway | IL2 Signaling Pathway | |||
| TCR Signaling Pathway | ||||
| Pathway Interaction Database | Regulation of nuclear SMAD2/3 signaling | |||
| Signaling events mediated by HDAC Class II | ||||
| FOXA2 and FOXA3 transcription factor networks | ||||
| Glucocorticoid receptor regulatory network | ||||
| Regulation of Androgen receptor activity | ||||
| AP-1 transcription factor network | ||||
| Reactome | BMAL1:CLOCK,NPAS2 activates circadian gene expression | |||
| WikiPathways | Serotonin Receptor 4/6/7 and NR3C Signaling | |||
| SIDS Susceptibility Pathways | ||||
| Nuclear Receptors Meta-Pathway | ||||
| Endoderm Differentiation | ||||
| Hair Follicle Development: Cytodifferentiation (Part 3 of 3) | ||||
| Adipogenesis | ||||
| Circadian Clock | ||||
| Nuclear Receptors | ||||
| References | Top | |||
|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 2805). | |||
| REF 2 | ClinicalTrials.gov (NCT02342002) Mifepristone and Misoprostol Versus Misoprostol Alone for Missed Abortion: A Randomized-controlled Trial. U.S. National Institutes of Health. | |||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||
| REF 4 | Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019 Jun;570(7762):462-467. | |||
| REF 5 | Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018 Mar 29;555(7698):623-628. | |||
| REF 6 | The glucocorticoid agonist activities of mifepristone (RU486) and progesterone are dependent on glucocorticoid receptor levels but not on EC50 values. Steroids. 2007 Jun;72(6-7):600-8. | |||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

