Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T02617
(Former ID: TTDI02373)
|
|||||
| Target Name |
Transferrin (TF)
|
|||||
| Synonyms |
Siderophilin; Serotransferrin; PRO1400; Beta-1 metal-binding globulin
Click to Show/Hide
|
|||||
| Gene Name |
TF
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
It is responsible for the transport of iron from sites of absorption and heme degradation to those of storage and utilization. Serum transferrin may also have a further role in stimulating cell proliferation. Transferrins are iron binding transport proteins which can bind two Fe(3+) ions in association with the binding of an anion, usually bicarbonate.
Click to Show/Hide
|
|||||
| BioChemical Class |
Transferrin
|
|||||
| UniProt ID | ||||||
| Sequence |
MRLAVGALLVCAVLGLCLAVPDKTVRWCAVSEHEATKCQSFRDHMKSVIPSDGPSVACVK
KASYLDCIRAIAANEADAVTLDAGLVYDAYLAPNNLKPVVAEFYGSKEDPQTFYYAVAVV KKDSGFQMNQLRGKKSCHTGLGRSAGWNIPIGLLYCDLPEPRKPLEKAVANFFSGSCAPC ADGTDFPQLCQLCPGCGCSTLNQYFGYSGAFKCLKDGAGDVAFVKHSTIFENLANKADRD QYELLCLDNTRKPVDEYKDCHLAQVPSHTVVARSMGGKEDLIWELLNQAQEHFGKDKSKE FQLFSSPHGKDLLFKDSAHGFLKVPPRMDAKMYLGYEYVTAIRNLREGTCPEAPTDECKP VKWCALSHHERLKCDEWSVNSVGKIECVSAETTEDCIAKIMNGEADAMSLDGGFVYIAGK CGLVPVLAENYNKSDNCEDTPEAGYFAIAVVKKSASDLTWDNLKGKKSCHTAVGRTAGWN IPMGLLYNKINHCRFDEFFSEGCAPGSKKDSSLCKLCMGSGLNLCEPNNKEGYYGYTGAF RCLVEKGDVAFVKHQTVPQNTGGKNPDPWAKNLNEKDYELLCLDGTRKPVEEYANCHLAR APNHAVVTRKDKEACVHKILRQQQHLFGSNVTDCSGNFCLFRSETKDLLFRDDTVCLAKL HDRNTYEKYLGEEYVKAVGNLRKCSTSSLLEACTFRRP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A00140 ; BADD_A02651 ; BADD_A05231 | |||||
| HIT2.0 ID | T20MBF | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | CALAA-01 | Drug Info | Phase 1 | Solid tumour/cancer | [2] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | EP-2167 | Drug Info | Preclinical | Solid tumour/cancer | [3] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | CALAA-01 | Drug Info | [1] | |||
| Inducer | [+] 1 Inducer drugs | + | ||||
| 1 | EP-2167 | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Nitrilotriacetic acid | Ligand Info | |||||
| Structure Description | Bismuth bound human serum transferrin | PDB:4H0W | ||||
| Method | X-ray diffraction | Resolution | 2.40 Å | Mutation | No | [5] |
| PDB Sequence |
VPDKTVRWCA
10 VSEHEATKCQ20 SFRDHMKSVI30 PSDGPSVACV40 KKASYLDCIR50 AIAANEADAV 60 TLDAGLVYDA70 YLAPNNLKPV80 VAEFYGSKED90 PQTFYYAVAV100 VKKDSGFQMN 110 QLRGKKSCHT120 GLGRSAGWNI130 PIGLLYCDLP140 EPRKPLEKAV150 ANFFSGSCAP 160 CADGTDFPQL170 CQLCPGCGCS180 TLNQYFGYSG190 AFKCLKDGAG200 DVAFVKHSTI 210 FENLANKADR220 DQYELLCLDN230 TRKPVDEYKD240 CHLAQVPSHT250 VVARSMGGKE 260 DLIWELLNQA270 QEHFGKDKSK280 EFQLFSSPHG290 KDLLFKDSAH300 GFLKVPPRMD 310 AKMYLGYEYV320 TAIRNLREGT330 CPEAPTDECK340 PVKWCALSHH350 ERLKCDEWSV 360 NSVGKIECVS370 AETTEDCIAK380 IMNGEADAMS390 LDGGFVYIAG400 KCGLVPVLAE 410 NYNKSDNCED420 TPEAGYFAIA430 VVKKSASDLT440 WDNLKGKKSC450 HTAVGRTAGW 460 NIPMGLLYNK470 INHCRFDEFF480 SEGCAPGSKK490 DSSLCKLCMG500 SGLNLCEPNN 510 KEGYYGYTGA520 FRCLVEKGDV530 AFVKHQTVPQ540 NTGGKNPDPW550 AKNLNEKDYE 560 LLCLDGTRKP570 VEEYANCHLA580 RAPNHAVVTR590 KDKEACVHKI600 LRQQQHLFGS 610 NVTDCSGNFC620 LFRSETKDLL630 FRDDTVCLAK640 LHDRNTYEKY650 LGEEYVKAVG 660 NLRKCSTSSL670 LEACTFRRP
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Pyrophosphate 2- | Ligand Info | |||||
| Structure Description | Crystal structure of human transferrin bound to Triferic FPC iron pyrophosphate | PDB:6CTC | ||||
| Method | X-ray diffraction | Resolution | 2.60 Å | Mutation | No | [6] |
| PDB Sequence |
DKTVRWCAVS
12 EHEATKCQSF22 RDHMKSVIPS32 DGPSVACVKK42 ASYLDCIRAI52 AANEADAVTL 62 DAGLVYDAYL72 APNNLKPVVA82 EFYGSKEDPQ92 TFYYAVAVVK102 KDSGFQMNQL 112 RGKKSCHTGL122 GRSAGWNIPI132 GLLYCDLPEP142 RKPLEKAVAN152 FFSGSCAPCA 162 DGTDFPQLCQ172 LCPGCGCSTL182 NQYFGYSGAF192 KCLKDGAGDV202 AFVKHSTIFE 212 NLANKADRDQ222 YELLCLDNTR232 KPVDEYKDCH242 LAQVPSHTVV252 ARSMGGKEDL 262 IWELLNQAQE272 HFGKDKSKEF282 QLFSSPHGKD292 LLFKDSAHGF302 LKVPPRMDAK 312 MYLGYEYVTA322 IRNLREGTCP332 EECKPVKWCA346 LSHHERLKCD356 EWSVNSVGKI 366 ECVSAETTED376 CIAKIMNGEA386 DAMSLDGGFV396 YIAGKCGLVP406 VLAENYNKSD 416 NCEDTPEAGY426 FAIAVVKKSA436 SDLTWDNLKG446 KKSCHTAVGR456 TAGWNIPMGL 466 LYNKINHCRF476 DEFFSEGCAP486 GSKKDSSLCK496 LCMGSGLNLC506 EPNNKEGYYG 516 YTGAFRCLVE526 KGDVAFVKHQ536 TVPQNTGGKN546 PDPWAKNLNE556 KDYELLCLDG 566 TRKPVEEYAN576 CHLARAPNHA586 VVTRKDKEAC596 VHKILRQQQH606 LFGSNVTDCS 616 GNFCLFRSET626 KDLLFRDDTV636 CLAKLHDRNT646 YEKYLGEEYV656 KAVGNLRKCS 666 TSSLLEACTF676 RRP
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
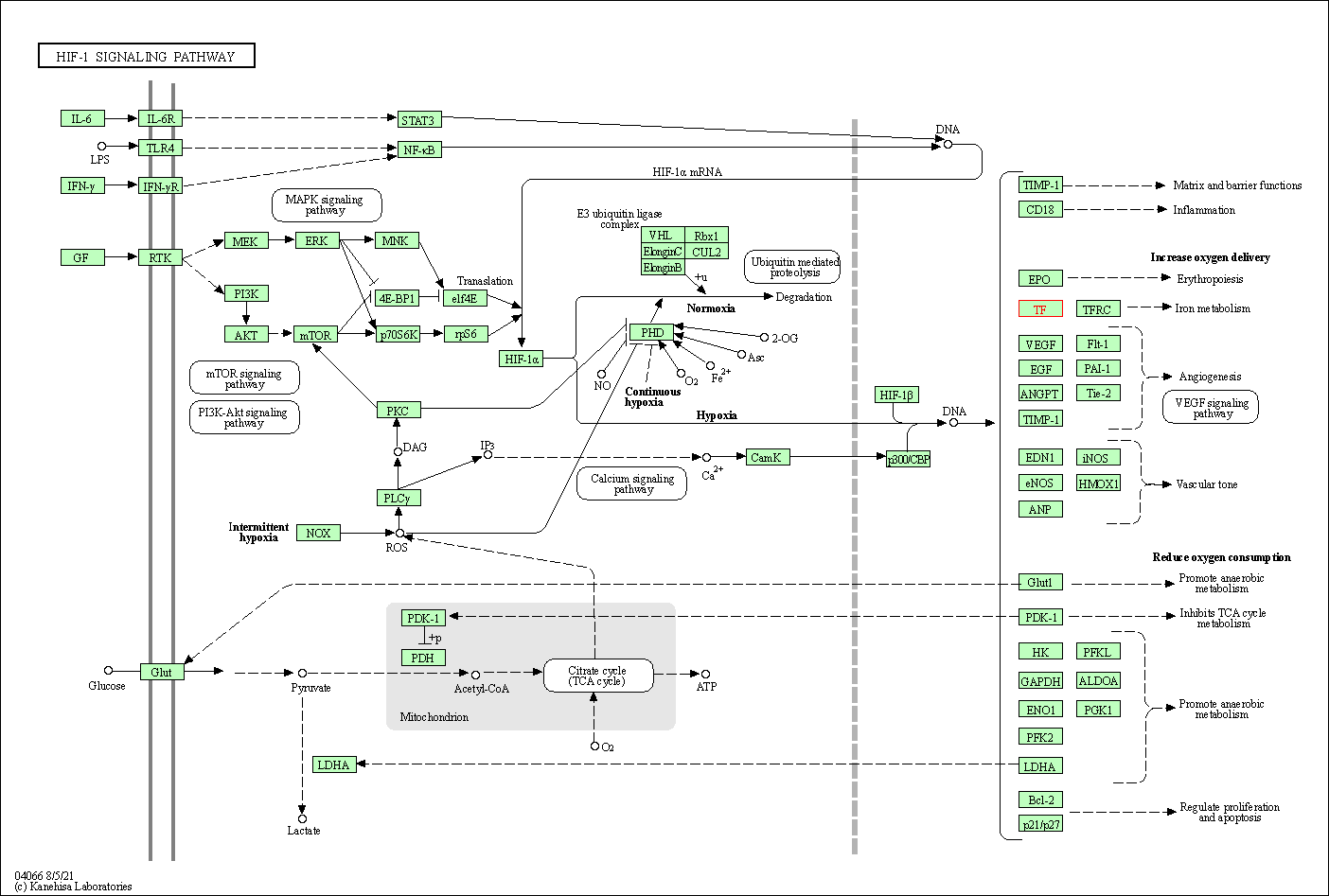
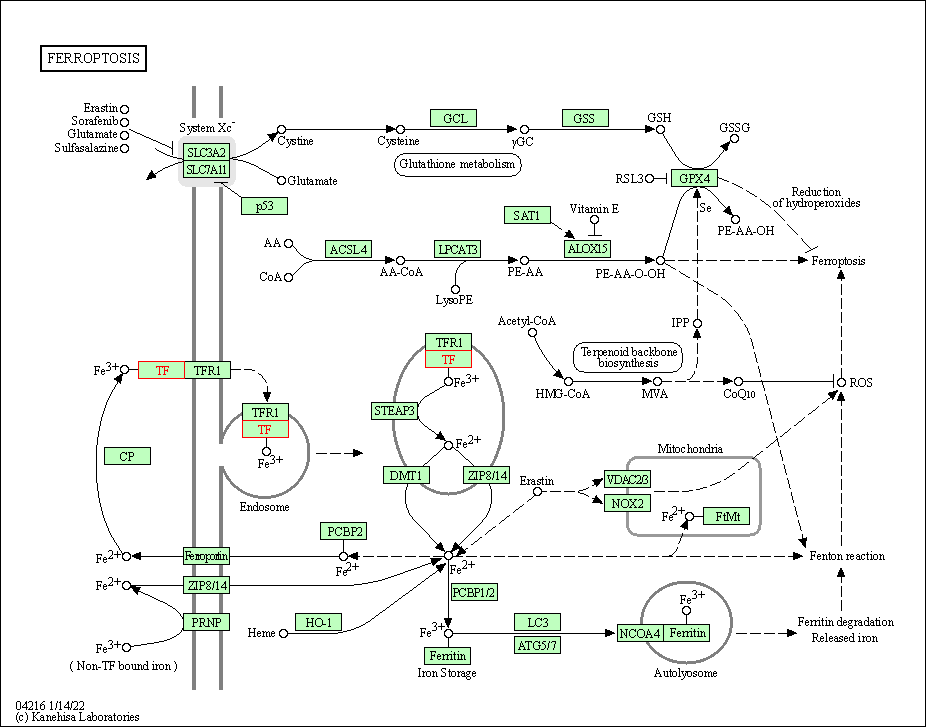
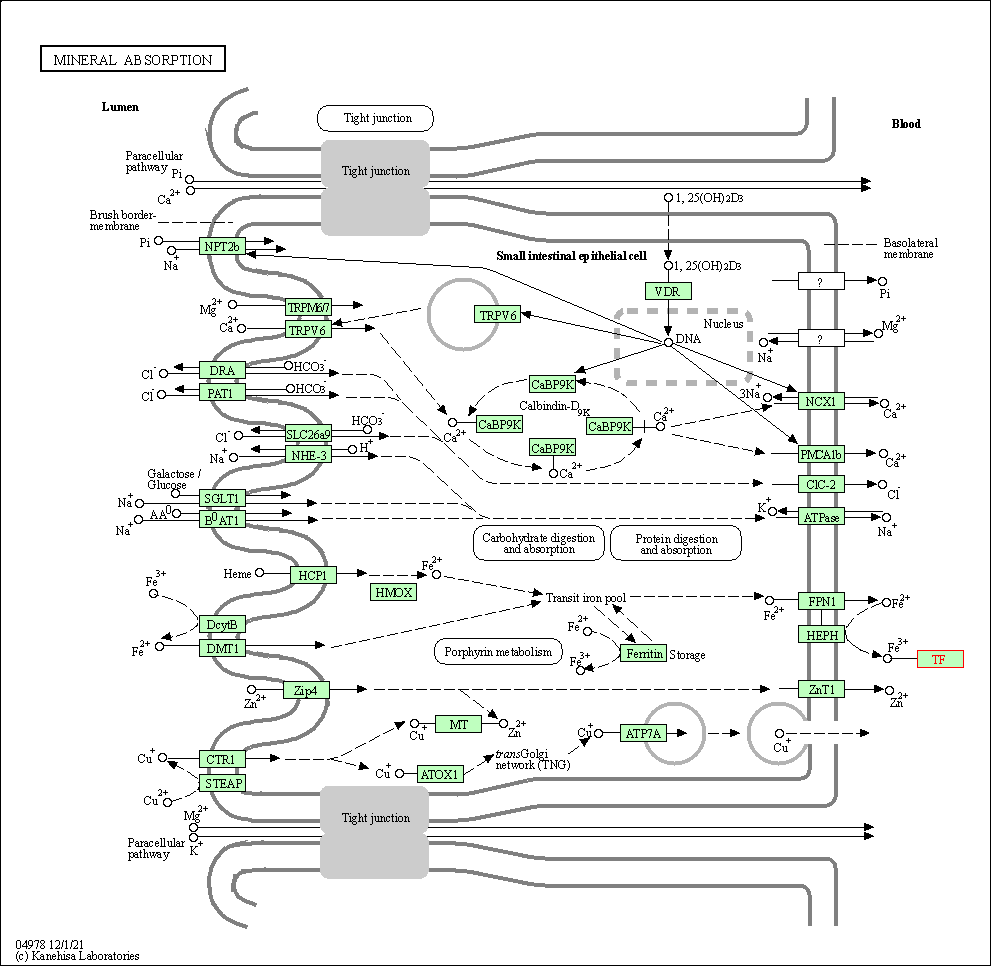
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ferroptosis | hsa04216 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Mineral absorption | hsa04978 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 1.54E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.82E-01 | Radiality | 1.30E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 2.00E+01 | Topological coefficient | 5.00E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-regulating Transcription Factors | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 2 KEGG Pathways | + | ||||
| 1 | HIF-1 signaling pathway | |||||
| 2 | Mineral absorption | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | TGF_beta_Receptor Signaling Pathway | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | EPHB forward signaling | |||||
| 2 | HIF-1-alpha transcription factor network | |||||
| Reactome | [+] 3 Reactome Pathways | + | ||||
| 1 | Platelet degranulation | |||||
| 2 | Iron uptake and transport | |||||
| 3 | Transferrin endocytosis and recycling | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | SIDS Susceptibility Pathways | |||||
| 2 | Differentiation Pathway | |||||
| 3 | Iron uptake and transport | |||||
| 4 | Iron metabolism in placenta | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 2 | ClinicalTrials.gov (NCT00689065) Safety Study of CALAA-01 to Treat Solid Tumor Cancers. U.S. National Institutes of Health. | |||||
| REF 3 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800017941) | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800017941) | |||||
| REF 5 | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe. Sci Rep. 2012;2:999. | |||||
| REF 6 | Ferric pyrophosphate citrate: interactions with transferrin. Biometals. 2018 Dec;31(6):1081-1089. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

