Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T15569
(Former ID: TTDS00482)
|
|||||
| Target Name |
Methylmalonyl-CoA mutase (MMUT)
|
|||||
| Synonyms |
Methylmalonyl-CoA isomerase; MUT; MCM
Click to Show/Hide
|
|||||
| Gene Name |
MUT
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Vitamin deficiency [ICD-11: 5B55-5B5F] | |||||
| Function |
Involved in the degradation of several amino acids, odd- chain fatty acids and cholesterol via propionyl-CoA to the tricarboxylic acid cycle. MCM has different functions in other species.
Click to Show/Hide
|
|||||
| BioChemical Class |
Intramolecular transferases
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 5.4.99.2
|
|||||
| Sequence |
MLRAKNQLFLLSPHYLRQVKESSGSRLIQQRLLHQQQPLHPEWAALAKKQLKGKNPEDLI
WHTPEGISIKPLYSKRDTMDLPEELPGVKPFTRGPYPTMYTFRPWTIRQYAGFSTVEESN KFYKDNIKAGQQGLSVAFDLATHRGYDSDNPRVRGDVGMAGVAIDTVEDTKILFDGIPLE KMSVSMTMNGAVIPVLANFIVTGEEQGVPKEKLTGTIQNDILKEFMVRNTYIFPPEPSMK IIADIFEYTAKHMPKFNSISISGYHMQEAGADAILELAYTLADGLEYSRTGLQAGLTIDE FAPRLSFFWGIGMNFYMEIAKMRAGRRLWAHLIEKMFQPKNSKSLLLRAHCQTSGWSLTE QDPYNNIVRTAIEAMAAVFGGTQSLHTNSFDEALGLPTVKSARIARNTQIIIQEESGIPK VADPWGGSYMMECLTNDVYDAALKLINEIEEMGGMAKAVAEGIPKLRIEECAARRQARID SGSEVIVGVNKYQLEKEDAVEVLAIDNTSVRNRQIEKLKKIKSSRDQALAERCLAALTEC AASGDGNILALAVDASRARCTVGEITDALKKVFGEHKANDRMVSGAYRQEFGESKEITSA IKRVHKFMEREGRRPRLLVAKMGQDGHDRGAKVIATGFADLGFDVDIGPLFQTPREVAQQ AVDADVHAVGISTLAAGHKTLVPELIKELNSLGRPDILVMCGGVIPPQDYEFLFEVGVSN VFGPGTRIPKAAVQVLDDIEKCLEKKQQSV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Hydroxocobalamin | Drug Info | Approved | Vitamin B12 deficiency | [2] | |
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | mRNA-3704 | Drug Info | Phase 1/2 | Methylmalonic acidemia | [3] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Cofactor | [+] 1 Cofactor drugs | + | ||||
| 1 | Hydroxocobalamin | Drug Info | [1] | |||
| Replacement | [+] 1 Replacement drugs | + | ||||
| 1 | mRNA-3704 | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Malonyl-CoA | Ligand Info | |||||
| Structure Description | Crystal structure of human methylmalonyl-CoA mutase in complex with adenosylcobalamin and malonyl-CoA | PDB:2XIQ | ||||
| Method | X-ray diffraction | Resolution | 1.95 Å | Mutation | No | [5] |
| PDB Sequence |
QQPLHPEWAA
45 LAKKQLKGKN55 PEDLIWHTPE65 GISIKPLYSK75 RDTMDLPEEL85 PGVKPFTRGP 95 YPTMYTFRPW105 TIRQYAGFST115 VEESNKFYKD125 NIKAGQQGLS135 VAFDLATHRG 145 YDSDNPRVRG155 DVGMAGVAID165 TVEDTKILFD175 GIPLEKMSVS185 MTMNGAVIPV 195 LANFIVTGEE205 QGVPKEKLTG215 TIQNDILKEF225 MVRNTYIFPP235 EPSMKIIADI 245 FEYTAKHMPK255 FNSISISGYH265 MQEAGADAIL275 ELAYTLADGL285 EYSRTGLQAG 295 LTIDEFAPRL305 SFFWGIGMNF315 YMEIAKMRAG325 RRLWAHLIEK335 MFQPKNSKSL 345 LLRAHCQTSG355 WSLTEQDPYN365 NIVRTAIEAM375 AAVFGGTQSL385 HTNSFDEALG 395 LPTVKSARIA405 RNTQIIIQEE415 SGIPKVADPW425 GGSYMMECLT435 NDVYDAALKL 445 INEIEEMGGM455 AKAVAEGIPK465 LRIEECAARR475 QARIDSGSEV485 IVGVNKYQLE 495 KEDTVEVLAI505 DNTSVRNRQI515 EKLKKIKSSR525 DQALAERCLA535 ALTECAASGD 545 GNILALAVDA555 SRARCTVGEI565 TDALKKVFGE575 HKANDRMVSG585 AYRQEFGESK 595 EITSAIKRVH605 KFMEREGRRP615 RLLVAKMGQD625 GHDRGAKVIA635 TGFADLGFDV 645 DIGPLFQTPR655 EVAQQAVDAD665 VHAVGVSTLA675 AGHKTLVPEL685 IKELNSLGRP 695 DILVMCGGVI705 PPQDYEFLFE715 VGVSNVFGPG725 TRIPKAAVQV735 LDDIEKCLEK 745 KQQS
|
|||||
|
|
TYR96
2.656
THR98
3.016
MET99
3.139
PHE102
3.727
ARG103
3.381
THR106
2.608
ARG108
2.822
TYR110
2.677
SER135
3.609
SER183
4.338
SER185
3.006
THR187
3.234
THR216
2.841
GLN218
3.136
ARG228
2.630
|
|||||
| Ligand Name: 5'-Deoxyadenosine | Ligand Info | |||||
| Structure Description | Crystal structure of human methylmalonyl-CoA mutase in complex with adenosylcobalamin | PDB:2XIJ | ||||
| Method | X-ray diffraction | Resolution | 1.95 Å | Mutation | Yes | [5] |
| PDB Sequence |
QQPLHPEWAA
45 LAKKQLKGKN55 PEDLIWHTPE65 GISIKPLYSK75 RDTMDLPEEL85 PGVKPFTRGP 95 YPTMYTFRPW105 TIRQYAGFST115 VEESNKFYKD125 NIKAGQQGLS135 VAFDLATHRG 145 YDSDNPRVRG155 DVGMAGVAID165 TVEDTKILFD175 GIPLEKMSVS185 MTMNGAVIPV 195 LANFIVTGEE205 QGVPKEKLTG215 TIQNDILKEF225 MVRNTYIFPP235 EPSMKIIADI 245 FEYTAKHMPK255 FNSISISGYH265 MQEAGADAIL275 ELAYTLADGL285 EYSRTGLQAG 295 LTIDEFAPRL305 SFFWGIGMNF315 YMEIAKMRAG325 RRLWAHLIEK335 MFQPKNSKSL 345 LLRAHCQTSG355 WSLTEQDPYN365 NIVRTAIEAM375 AAVFGGTQSL385 HTNSFDEALG 395 LPTVKSARIA405 RNTQIIIQEE415 SGIPKVADPW425 GGSYMMECLT435 NDVYDAALKL 445 INEIEEMGGM455 AKAVAEGIPK465 LRIEECAARR475 QARIDSGSEV485 IVGVNKYQLE 495 KEDTVEVLAI505 DNTSVRNRQI515 EKLKKIKSSR525 DQALAERCLA535 ALTECAASGD 545 GNILALAVDA555 SRARCTVGEI565 TDALKKVFGE575 HKANDRMVSG585 AYRQEFGESK 595 EITSAIKRVH605 KFMEREGRRP615 RLLVAKMGQD625 GHDRGAKVIA635 TGFADLGFDV 645 DIGPLFQTPR655 EVAQQAVDAD665 VHAVGVSTLA675 AGHKTLVPEL685 IKELNSLGRP 695 DILVMCGGVI705 PPQDYEFLFE715 VGVSNVFGPG725 TRIPKAAVQV735 LDDIEKCLEK 745
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Valine, leucine and isoleucine degradation | hsa00280 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Glyoxylate and dicarboxylate metabolism | hsa00630 | Affiliated Target |

|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Propanoate metabolism | hsa00640 | Affiliated Target |
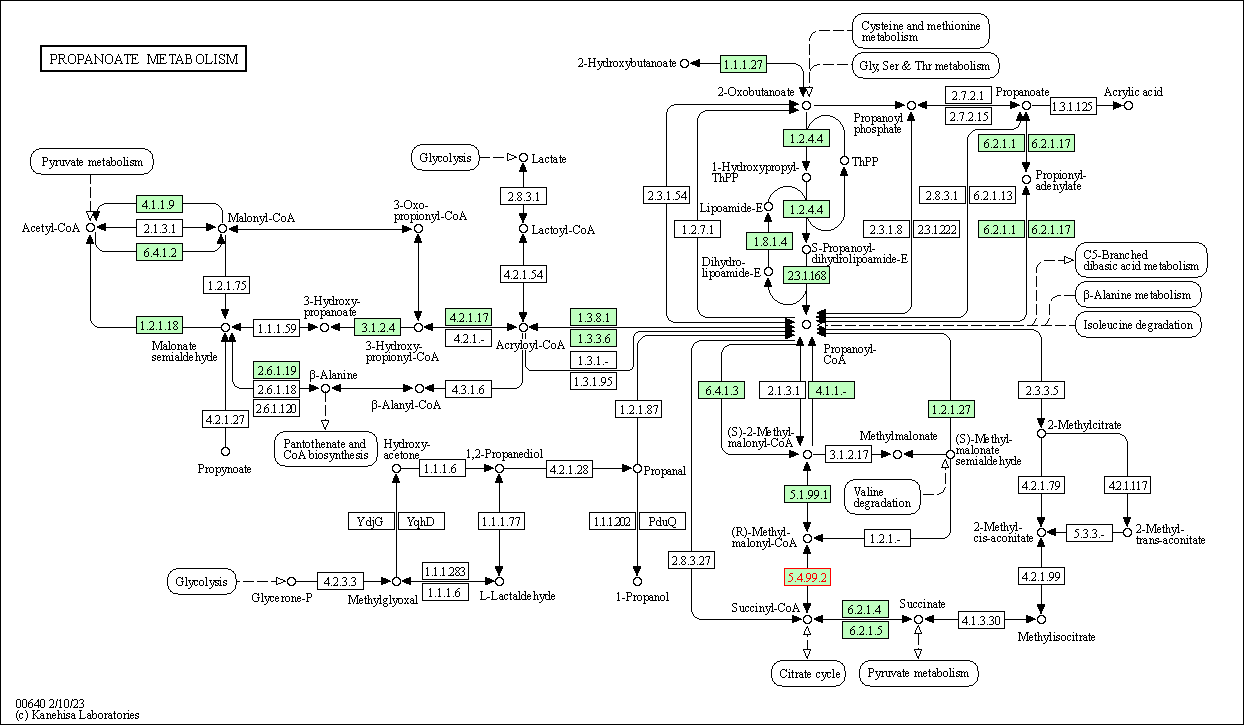
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 6.17E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.79E-01 | Radiality | 1.29E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 8.00E+00 | Topological coefficient | 2.52E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 3 BioCyc Pathways | + | ||||
| 1 | Superpathway of methionine degradation | |||||
| 2 | Propionyl-CoA degradation | |||||
| 3 | 2-oxobutanoate degradation | |||||
| KEGG Pathway | [+] 5 KEGG Pathways | + | ||||
| 1 | Valine, leucine and isoleucine degradation | |||||
| 2 | Glyoxylate and dicarboxylate metabolism | |||||
| 3 | Propanoate metabolism | |||||
| 4 | Metabolic pathways | |||||
| 5 | Carbon metabolism | |||||
| Panther Pathway | [+] 2 Panther Pathways | + | ||||
| 1 | Methylmalonyl pathway | |||||
| 2 | Succinate to proprionate conversion | |||||
| Pathwhiz Pathway | [+] 3 Pathwhiz Pathways | + | ||||
| 1 | Valine, Leucine and Isoleucine Degradation | |||||
| 2 | Propanoate Metabolism | |||||
| 3 | Threonine and 2-Oxobutanoate Degradation | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Cobalamin (Cbl, vitamin B12) transport and metabolism | |||||
| WikiPathways | [+] 3 WikiPathways | + | ||||
| 1 | Metabolism of water-soluble vitamins and cofactors | |||||
| 2 | Fatty acid, triacylglycerol, and ketone body metabolism | |||||
| 3 | Vitamin B12 Metabolism | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Vitamin B12. Nippon Rinsho. 1999 Oct;57(10):2205-10. | |||||
| REF 2 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (ANDA) 085998. | |||||
| REF 3 | ClinicalTrials.gov (NCT03810690) Open Label Study of mRNA-3704 in Patients With Isolated Methylmalonic Acidemia. U.S. National Institutes of Health. | |||||
| REF 4 | Clinical pipeline report, company report or official report of Moderna Therapeutics. | |||||
| REF 5 | Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation. J Biol Chem. 2010 Dec 3;285(49):38204-13. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

