Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T15823
(Former ID: TTDR01106)
|
|||||
| Target Name |
Breast cancer type 2 susceptibility protein (BRCA2)
|
|||||
| Synonyms |
Fanconi anemia group D1 protein; FANCD1; FACD
Click to Show/Hide
|
|||||
| Gene Name |
BRCA2
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Binds RAD51 and potentiates recombinational DNA repair by promoting assembly of RAD51 onto single-stranded DNA (ssDNA). Acts by targeting RAD51 to ssDNA over double-stranded DNA, enabling RAD51 to displace replication protein-A (RPA) from ssDNA and stabilizing RAD51-ssDNA filaments by blocking ATP hydrolysis. Part of a PALB2-scaffolded HR complex containing RAD51C and which is thought to play a role in DNA repair by HR. May participate in S phase checkpoint activation. Binds selectively to ssDNA, and to ssDNA in tailed duplexes and replication fork structures. May play a role in the extension step after strand invasion at replication-dependent DNA double-strand breaks; together with PALB2 is involved in both POLH localization at collapsed replication forks and DNA polymerization activity. In concert with NPM1, regulates centrosome duplication. Interacts with the TREX-2 complex (transcription and export complex 2) subunits PCID2 and SEM1, and is required to prevent R-loop-associated DNA damage and thus transcription-associated genomic instability. Silencing of BRCA2 promotes R-loop accumulation at actively transcribed genes in replicating and non-replicating cells, suggesting that BRCA2 mediates the control of R-loop associated genomic instability, independently of its known role in homologous recombination. Involved in double-strand break repair and/or homologous recombination.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MPIGSKERPTFFEIFKTRCNKADLGPISLNWFEELSSEAPPYNSEPAEESEHKNNNYEPN
LFKTPQRKPSYNQLASTPIIFKEQGLTLPLYQSPVKELDKFKLDLGRNVPNSRHKSLRTV KTKMDQADDVSCPLLNSCLSESPVVLQCTHVTPQRDKSVVCGSLFHTPKFVKGRQTPKHI SESLGAEVDPDMSWSSSLATPPTLSSTVLIVRNEEASETVFPHDTTANVKSYFSNHDESL KKNDRFIASVTDSENTNQREAASHGFGKTSGNSFKVNSCKDHIGKSMPNVLEDEVYETVV DTSEEDSFSLCFSKCRTKNLQKVRTSKTRKKIFHEANADECEKSKNQVKEKYSFVSEVEP NDTDPLDSNVANQKPFESGSDKISKEVVPSLACEWSQLTLSGLNGAQMEKIPLLHISSCD QNISEKDLLDTENKRKKDFLTSENSLPRISSLPKSEKPLNEETVVNKRDEEQHLESHTDC ILAVKQAISGTSPVASSFQGIKKSIFRIRESPKETFNASFSGHMTDPNFKKETEASESGL EIHTVCSQKEDSLCPNLIDNGSWPATTTQNSVALKNAGLISTLKKKTNKFIYAIHDETSY KGKKIPKDQKSELINCSAQFEANAFEAPLTFANADSGLLHSSVKRSCSQNDSEEPTLSLT SSFGTILRKCSRNETCSNNTVISQDLDYKEAKCNKEKLQLFITPEADSLSCLQEGQCEND PKSKKVSDIKEEVLAAACHPVQHSKVEYSDTDFQSQKSLLYDHENASTLILTPTSKDVLS NLVMISRGKESYKMSDKLKGNNYESDVELTKNIPMEKNQDVCALNENYKNVELLPPEKYM RVASPSRKVQFNQNTNLRVIQKNQEETTSISKITVNPDSEELFSDNENNFVFQVANERNN LALGNTKELHETDLTCVNEPIFKNSTMVLYGDTGDKQATQVSIKKDLVYVLAEENKNSVK QHIKMTLGQDLKSDISLNIDKIPEKNNDYMNKWAGLLGPISNHSFGGSFRTASNKEIKLS EHNIKKSKMFFKDIEEQYPTSLACVEIVNTLALDNQKKLSKPQSINTVSAHLQSSVVVSD CKNSHITPQMLFSKQDFNSNHNLTPSQKAEITELSTILEESGSQFEFTQFRKPSYILQKS TFEVPENQMTILKTTSEECRDADLHVIMNAPSIGQVDSSKQFEGTVEIKRKFAGLLKNDC NKSASGYLTDENEVGFRGFYSAHGTKLNVSTEALQKAVKLFSDIENISEETSAEVHPISL SSSKCHDSVVSMFKIENHNDKTVSEKNNKCQLILQNNIEMTTGTFVEEITENYKRNTENE DNKYTAASRNSHNLEFDGSDSSKNDTVCIHKDETDLLFTDQHNICLKLSGQFMKEGNTQI KEDLSDLTFLEVAKAQEACHGNTSNKEQLTATKTEQNIKDFETSDTFFQTASGKNISVAK ESFNKIVNFFDQKPEELHNFSLNSELHSDIRKNKMDILSYEETDIVKHKILKESVPVGTG NQLVTFQGQPERDEKIKEPTLLGFHTASGKKVKIAKESLDKVKNLFDEKEQGTSEITSFS HQWAKTLKYREACKDLELACETIEITAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNL CRQTENLKTSKSIFLKVKVHENVEKETAKSPATCYTNQSPYSVIENSALAFYTSCSRKTS VSQTSLLEAKKWLREGIFDGQPERINTADYVGNYLYENNSNSTIAENDKNHLSEKQDTYL SNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVKDANAY PQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIK KVKDIFTDSFSKVIKENNENKSKICQTKIMAGCYEALDDSEDILHNSLDNDECSTHSHKV FADIQSEEILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFST ASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLI SQKGFSYNVVNSSAFSGFSTASGKQVSILESSLHKVKGVLEEFDLIRTEHSLHYSPTSRQ NVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSENNHSIKVSPYLSQFQQ DKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTYSKDSE NYFETEAVEIAKAFMEDDELTDSKLPSHATHSLFTCPENEEMVLSNSRIGKRRGEPLILV GEPSIKRNLLNEFDRIIENQEKSLKASKSTPDGTIKDRRLFMHHVSLEPITCVPFRTTKE RQEIQNPNFTAPGQEFLSKSHLYEHLTLEKSSSNLAVSGHPFYQVSATRNEKMRHLITTG RPTKVFVPPFKTKSHFHRVEQCVRNINLEENRQKQNIDGHGSDDSKNKINDNEIHQFNKN NSNQAAAVTFTKCEEEPLDLITSLQNARDIQDMRIKKKQRQRVFPQPGSLYLAKTSTLPR ISLKAAVGGQVPSACSHKQLYTYGVSKHCIKINSKNAESFQFHTEDYFGKESLWTGKGIQ LADGGWLIPSNDGKAGKEEFYRALCDTPGVDPKLISRIWVYNHYRWIIWKLAAMECAFPK EFANRCLSPERVLLQLKYRYDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANI SETSSNKTSSADTQKVAIIELTDGWYAVKAQLDPPLLAVLKNGRLTVGQKIILHGAELVG SPDACTPLEAPESLMLKISANSTRPARWYTKLGFFPDPRPFPLPLSSLFSDGGNVGCVDV IIQRAYPIQWMEKTSSGLYIFRNEREEEKEAAKYVEAQQKRLEALFTKIQEEFEEHEENT TKPYLPSRALTRQQVRALQDGAELYEAVKNAADPAYLEGYFSEEQLRALNNHRQMLNDKK QAQIQLEIRKAMESAEQKEQGLSRDVTTVWKLRIVSYSKKEKDSVILSIWRPSSDLYSLL TEGKRYRIYHLATSKSKSKSERANIQLAATKKTQYQQLPVSDEILFQIYQPREPLHFSKF LDPDFQPSCSEVDLIGFVVSVVKKTGLAPFVYLSDECYNLLAIKFWIDLNEDIIKPHMLI AASNLQWRPESKSGLLTLFAGDFSVFSASPKEGHFQETFNKMKNTVENIDILCNEAENKL MHILHANDPKWSTPTKDCTSGPYTAQIIPGTGNKLLMSSPNCEIYYQSPLSLCMAKRKSV STPVSAQMTSKSCKGEKEIDDQKNCKKRRALDFLSRLPLPPPVSPICTFVSPAAQKAFQP PRSCGTKYETPIKKKELNSPQMTPFKKFNEISLLESNSIADEELALINTQALLSGSTGEK QFISVSESTRTAPTSSEDYLRLKRRCTTSLIKEQESSQASTEECEKNKQDTITTKKYI Click to Show/Hide
|
|||||
| HIT2.0 ID | T59EBF | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|

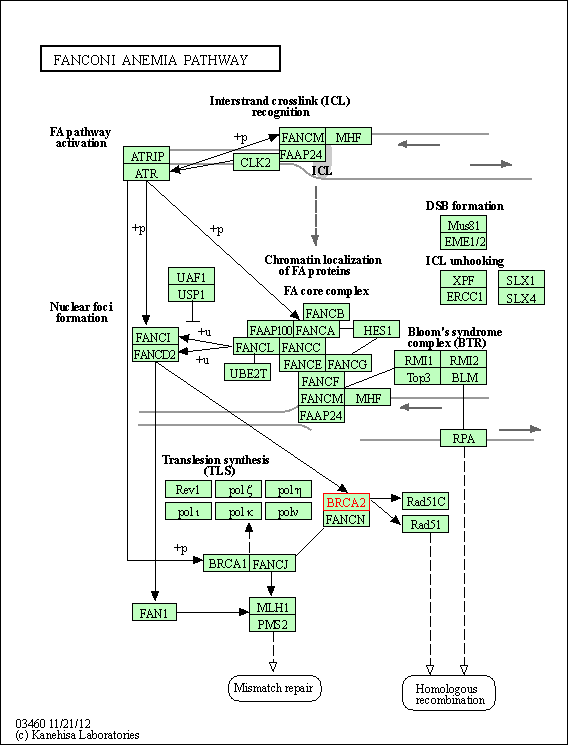
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Homologous recombination | hsa03440 | Affiliated Target |

|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
| Fanconi anemia pathway | hsa03460 | Affiliated Target |

|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
| Degree | 40 | Degree centrality | 4.30E-03 | Betweenness centrality | 1.74E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.42E-01 | Radiality | 1.42E+01 | Clustering coefficient | 1.72E-01 |
| Neighborhood connectivity | 3.52E+01 | Topological coefficient | 5.57E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Drug Resistance Mutation (DRM) |
||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Molecular prescreening to select patient population in early clinical trials. Nat Rev Clin Oncol. 2012 Apr 3;9(6):359-66. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

