Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T16514
(Former ID: TTDS00295)
|
|||||
| Target Name |
Fatty acid synthase (FASN)
|
|||||
| Synonyms |
Yeast fatty acid synthase; Fatty-acyl-CoA synthase; Fatty acyl-CoA synthetase enzyme; FAS
Click to Show/Hide
|
|||||
| Gene Name |
FASN
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | concerning food/fluid intake symptom [ICD-11: MG43] | |||||
| Function |
Fatty acid synthetase catalyzes the formation of long-chain fatty acids from acetyl-CoA, malonyl-CoA and NADPH. This multifunctional protein has 7 catalytic activities as an acyl carrier protein.
Click to Show/Hide
|
|||||
| BioChemical Class |
Acyltransferase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.3.1.85
|
|||||
| Sequence |
MEEVVIAGMSGKLPESENLQEFWDNLIGGVDMVTDDDRRWKAGLYGLPRRSGKLKDLSRF
DASFFGVHPKQAHTMDPQLRLLLEVTYEAIVDGGINPDSLRGTHTGVWVGVSGSETSEAL SRDPETLVGYSMVGCQRAMMANRLSFFFDFRGPSIALDTACSSSLMALQNAYQAIHSGQC PAAIVGGINVLLKPNTSVQFLRLGMLSPEGTCKAFDTAGNGYCRSEGVVAVLLTKKSLAR RVYATILNAGTNTDGFKEQGVTFPSGDIQEQLIRSLYQSAGVAPESFEYIEAHGTGTKVG DPQELNGITRALCATRQEPLLIGSTKSNMGHPEPASGLAALAKVLLSLEHGLWAPNLHFH SPNPEIPALLDGRLQVVDQPLPVRGGNVGINSFGFGGSNVHIILRPNTQPPPAPAPHATL PRLLRASGRTPEAVQKLLEQGLRHSQDLAFLSMLNDIAAVPATAMPFRGYAVLGGERGGP EVQQVPAGERPLWFICSGMGTQWRGMGLSLMRLDRFRDSILRSDEAVKPFGLKVSQLLLS TDESTFDDIVHSFVSLTAIQIGLIDLLSCMGLRPDGIVGHSLGEVACGYADGCLSQEEAV LAAYWRGQCIKEAHLPPGAMAAVGLSWEECKQRCPPGVVPACHNSKDTVTISGPQAPVFE FVEQLRKEGVFAKEVRTGGMAFHSYFMEAIAPPLLQELKKVIREPKPRSARWLSTSIPEA QWHSSLARTSSAEYNVNNLVSPVLFQEALWHVPEHAVVLEIAPHALLQAVLKRGLKPSCT IIPLMKKDHRDNLEFFLAGIGRLHLSGIDANPNALFPPVEFPAPRGTPLISPLIKWDHSL AWDVPAAEDFPNGSGSPSAAIYNIDTSSESPDHYLVDHTLDGRVLFPATGYLSIVWKTLA RALGLGVEQLPVVFEDVVLHQATILPKTGTVSLEVRLLEASRAFEVSENGNLVVSGKVYQ WDDPDPRLFDHPESPTPNPTEPLFLAQAEVYKELRLRGYDYGPHFQGILEASLEGDSGRL LWKDNWVSFMDTMLQMSILGSAKHGLYLPTRVTAIHIDPATHRQKLYTLQDKAQVADVVV SRWLRVTVAGGVHISGLHTESAPRRQQEQQVPILEKFCFTPHTEEGCLSERAALQEELQL CKGLVQALQTKVTQQGLKMVVPGLDGAQIPRDPSQQELPRLLSAACRLQLNGNLQLELAQ VLAQERPKLPEDPLLSGLLDSPALKACLDTAVENMPSLKMKVVEVLAGHGHLYSRIPGLL SPHPLLQLSYTATDRHPQALEAAQAELQQHDVAQGQWDPADPAPSALGSADLLVCNCAVA ALGDPASALSNMVAALREGGFLLLHTLLRGHPLGDIVAFLTSTEPQYGQGILSQDAWESL FSRVSLRLVGLKKSFYGSTLFLCRRPTPQDSPIFLPVDDTSFRWVESLKGILADEDSSRP VWLKAINCATSGVVGLVNCLRREPGGNRLRCVLLSNLSSTSHVPEVDPGSAELQKVLQGD LVMNVYRDGAWGAFRHFLLEEDKPEEPTAHAFVSTLTRGDLSSIRWVCSSLRHAQPTCPG AQLCTVYYASLNFRDIMLATGKLSPDAIPGKWTSQDSLLGMEFSGRDASGKRVMGLVPAK GLATSVLLSPDFLWDVPSNWTLEEAASVPVVYSTAYYALVVRGRVRPGETLLIHSGSGGV GQAAIAIALSLGCRVFTTVGSAEKRAYLQARFPQLDSTSFANSRDTSFEQHVLWHTGGKG VDLVLNSLAEEKLQASVRCLATHGRFLEIGKFDLSQNHPLGMAIFLKNVTFHGVLLDAFF NESSADWREVWALVQAGIRDGVVRPLKCTVFHGAQVEDAFRYMAQGKHIGKVVVQVLAEE PEAVLKGAKPKLMSAISKTFCPAHKSYIIAGGLGGFGLELAQWLIQRGVQKLVLTSRSGI RTGYQAKQVRRWRRQGVQVQVSTSNISSLEGARGLIAEAAQLGPVGGVFNLAVVLRDGLL ENQTPEFFQDVCKPKYSGTLNLDRVTREACPELDYFVVFSSVSCGRGNAGQSNYGFANSA MERICEKRRHEGLPGLAVQWGAIGDVGILVETMSTNDTIVSGTLPQRMASCLEVLDLFLN QPHMVLSSFVLAEKAAAYRDRDSQRDLVEAVAHILGIRDLAAVNLDSSLADLGLDSLMSV EVRQTLERELNLVLSVREVRQLTLRKLQELSSKADEASELACPTPKEDGLAQQQTQLNLR SLLVNPEGPTLMRLNSVQSSERPLFLVHPIEGSTTVFHSLASRLSIPTYGLQCTRAAPLD SIHSLAAYYIDCIRQVQPEGPYRVAGYSYGACVAFEMCSQLQAQQSPAPTHNSLFLFDGS PTYVLAYTQSYRAKLTPGCEAEAETEAICFFVQQFTDMEHNRVLEALLPLKGLEERVAAA VDLIIKSHQGLDRQELSFAARSFYYKLRAAEQYTPKAKYHGNVMLLRAKTGGAYGEDLGA DYNLSQVCDGKVSVHVIEGDHRTLLEGSGLESIISIIHSSLAEPRVSVREG Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A01285 | |||||
| HIT2.0 ID | T74JJN | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Cerulenin | Drug Info | Approved | Weight loss | [2] | |
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | Epigallocatechin gallate | Drug Info | Phase 3 | Hepatic fibrosis | [3], [4], [5], [6] | |
| 2 | TVB-2640 | Drug Info | Phase 2 | Astrocytoma | [7], [8] | |
| 3 | TVB-2640 | Drug Info | Phase 2 | Non-alcoholic fatty liver disease | [9] | |
| Preclinical Drug(s) | [+] 2 Preclinical Drugs | + | ||||
| 1 | FAS1 | Drug Info | Preclinical | Fungal infection | [10] | |
| 2 | FSA2 | Drug Info | Preclinical | Fungal infection | [10] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 19 Inhibitor drugs | + | ||||
| 1 | Cerulenin | Drug Info | [1], [11], [12] | |||
| 2 | Epigallocatechin gallate | Drug Info | [13] | |||
| 3 | TVB-2640 | Drug Info | [14] | |||
| 4 | TVB-2640 | Drug Info | [15] | |||
| 5 | FAS1 | Drug Info | [16] | |||
| 6 | FSA2 | Drug Info | [16] | |||
| 7 | (-)-CATECHINGALLATE | Drug Info | [13] | |||
| 8 | 2-Hexadecynoic acid | Drug Info | [17] | |||
| 9 | 3,7,3',4'-TETRAHYDROXYFLAVONE | Drug Info | [13] | |||
| 10 | 4-hydroxy-6-nitro-3-phenylquinolin-2(1H)-one | Drug Info | [18] | |||
| 11 | 4-hydroxy-8-nitro-3-phenylquinolin-2(1H)-one | Drug Info | [18] | |||
| 12 | biochanin A | Drug Info | [13] | |||
| 13 | C75 | Drug Info | [20] | |||
| 14 | FAS-031 | Drug Info | [19] | |||
| 15 | GALLOCATECHIN GALLATE | Drug Info | [13] | |||
| 16 | GSK2194069 | Drug Info | [21] | |||
| 17 | MG-28 | Drug Info | [19] | |||
| 18 | MORIN | Drug Info | [13] | |||
| 19 | MT-061 | Drug Info | [19] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: NADPH | Ligand Info | |||||
| Structure Description | Human Fatty Acid Synthase Psi/KR Tri-Domain with NADPH and Compound 22 | PDB:6NNA | ||||
| Method | X-ray diffraction | Resolution | 2.26 Å | Mutation | Yes | [22] |
| PDB Sequence |
QQVPILEKFC
1118 FTPHTEEGCL1128 SERAALQEEL1138 QLCKGLVQAL1148 QTPSQQELPR1180 LLSAACRLAQ 1200 VLAQERPKLP1210 EDPLLSGLLD1220 SPALKACLDT1230 AVENMPSLKM1240 KVVEVLAGHG 1250 HLYSRIPGLL1260 SPHPLLQLSY1270 TATDRHPQAL1280 EAAQAELQQH1290 DVAQGQWDPA 1300 DPAPSALGSA1310 DLLVCNCAVA1320 ALGDPASALS1330 NMVAALREGG1340 FLLLHTLLRG 1350 HPLGDIVAFL1360 TSQGILSQDA1376 WESLFSRVSL1386 RLVGLKKSFY1396 GSTLFLCRRP 1406 TPQDSPIFLP1416 VDDTSFRWVE1426 SLKGILADED1436 SARPVWLKAI1446 NCATSGVVGL 1456 VNCLRREPGG1466 NRLRCVLLSN1476 LSSTSHVPEV1486 DPGSAELQKV1496 LQGDLVMNVY 1506 RDGAWGAFRH1516 FLLEEDSKTF1880 PAHKSYIIAG1891 GLGGFGLELA1901 QWLIQRGVQK 1911 LVLTSRSGIR1921 TGYQAKQVRR1931 WRRQGVQVQV1941 STSNISSLEG1951 ARGLIAEAAQ 1961 LGPVGGVFNL1971 AVVLRDGLLE1981 NQTPEFFQDV1991 CKPKYSGTLN2001 LDRVTREACP 2011 ELDYFVVFSS2021 VSCGRGNAGQ2031 SNYGFANSAM2041 ERICEKRRHE2051 GLPGLAVQWG 2061 AIGDVGILVE2071 DTIVSGTLPQ2086 RMASCLEVLD2096 LFLNQPHMVL2106 SSFVLAE |
|||||
|
|
ALA1890
4.647
GLY1891
3.536
LEU1893
4.429
GLY1894
3.198
GLY1895
2.942
PHE1896
2.908
GLY1897
4.239
THR1915
3.284
SER1916
2.684
ARG1917
2.804
SER1918
3.472
GLY1919
4.616
ARG1921
4.104
SER1944
4.829
ASN1945
2.950
ILE1946
4.522
SER1947
3.684
ASN1970
4.280
LEU1971
3.412
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: GSK2194069 | Ligand Info | |||||
| Structure Description | Human Fatty Acid Synthase Psi/KR Tri-Domain with NADPH and GSK2194069 | PDB:4PIV | ||||
| Method | X-ray diffraction | Resolution | 2.30 Å | Mutation | Yes | [21] |
| PDB Sequence |
MQVPILEKFC
1118 FTPHTEEGCL1128 SERAALQEEL1138 QLCKGLVQAL1148 QTPSQQELPR1180 LLSAACRLAQ 1200 VLAQERPKLP1210 EDPLLSGLLD1220 SPALKACLDT1230 AVENMPSLKM1240 KVVEVLAGHG 1250 HLYSRIPGLL1260 SPHPLLQLSY1270 TATDRHPQAL1280 EAAQAELQQH1290 DVAQGQWDPA 1300 DPAPSALGSA1310 DLLVCNCAVA1320 ALGDPASALS1330 NMVAALREGG1340 FLLLHTLLRG 1350 HPLGDIVAFL1360 TSQGILSQDA1376 WESLFSRVSL1386 RLVGLKKSFY1396 GSTLFLCRRP 1406 TPQDSPIFLP1416 VDDTSFRWVE1426 SLKGILADED1436 SARPVWLKAI1446 NCATSGVVGL 1456 VNCLRREPGG1466 NRLRCVLLSN1476 LSSTSHVPEV1486 DPGSAELQKV1496 LQGDLVMNVY 1506 RDGAWGAFRH1516 FLLEEDSKTF1880 CPAHKSYIIA1890 GGLGGFGLEL1900 AQWLIQRGVQ 1910 KLVLTSRSGI1920 RTGYQAKQVR1930 RWRRQGVQVQ1940 VSTSNISSLE1950 GARGLIAEAA 1960 QLGPVGGVFN1970 LAVVLRDGLL1980 ENQTPEFFQD1990 VCKPKYSGTL2000 NLDRVTREAC 2010 PELDYFVVFS2020 SVSCGRGNAG2030 QSNYGFANSA2040 MERICEKRRH2050 EGLPGLAVQW 2060 GAIGDVGILV2070 ETNDTIVSGT2083 LPQRMASCLE2093 VLDLFLNQPH2103 MVLSSFVLAE 2113
|
|||||
|
|
HIS1263
3.383
PRO1264
3.349
LEU1265
4.112
LEU1975
3.690
SER2021
2.787
VAL2022
3.727
SER2023
3.305
ARG2026
3.489
GLY2027
3.716
ASN2028
3.132
GLN2031
3.884
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
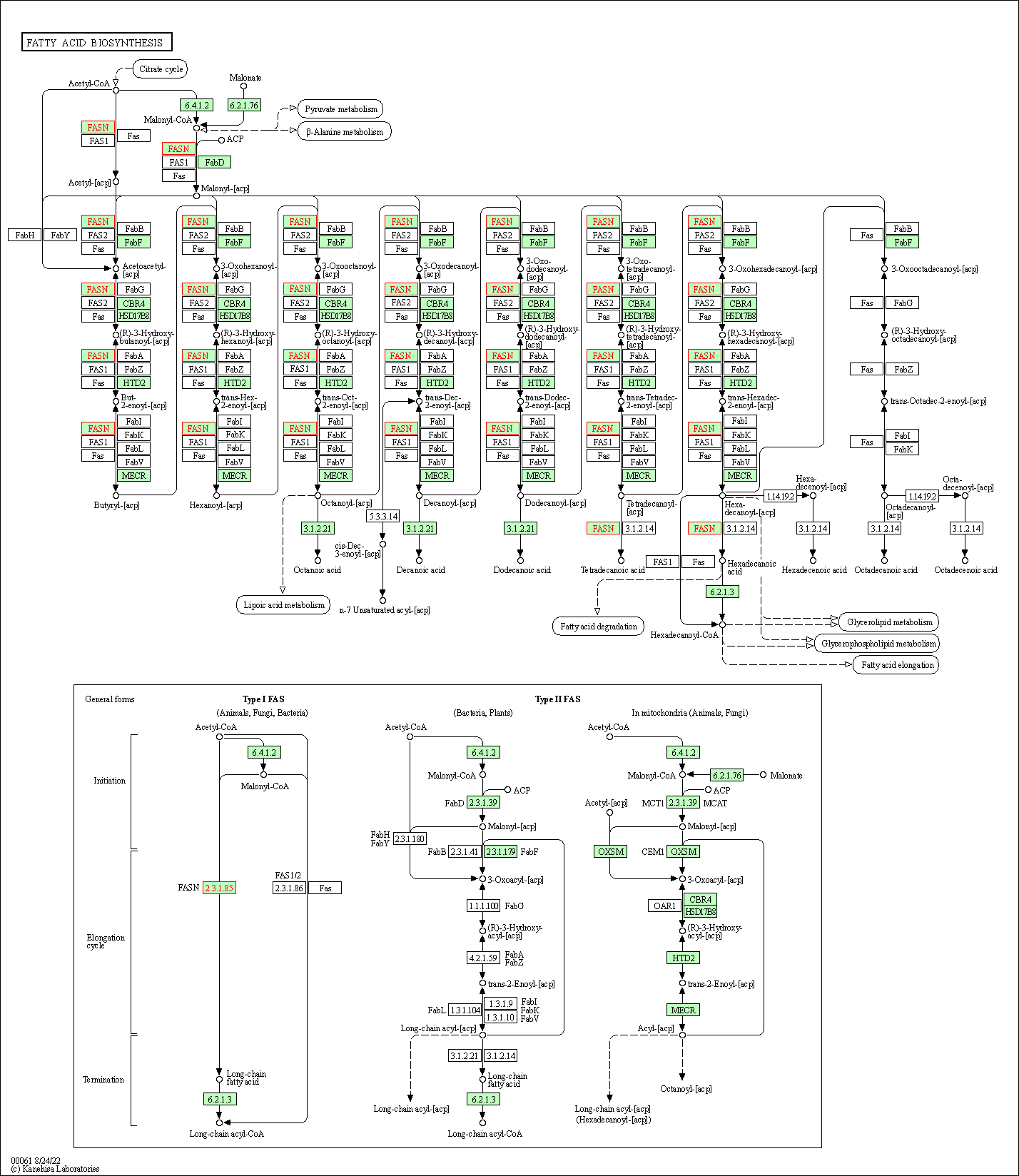
| Fatty acid biosynthesis | hsa00061 | Affiliated Target |

|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
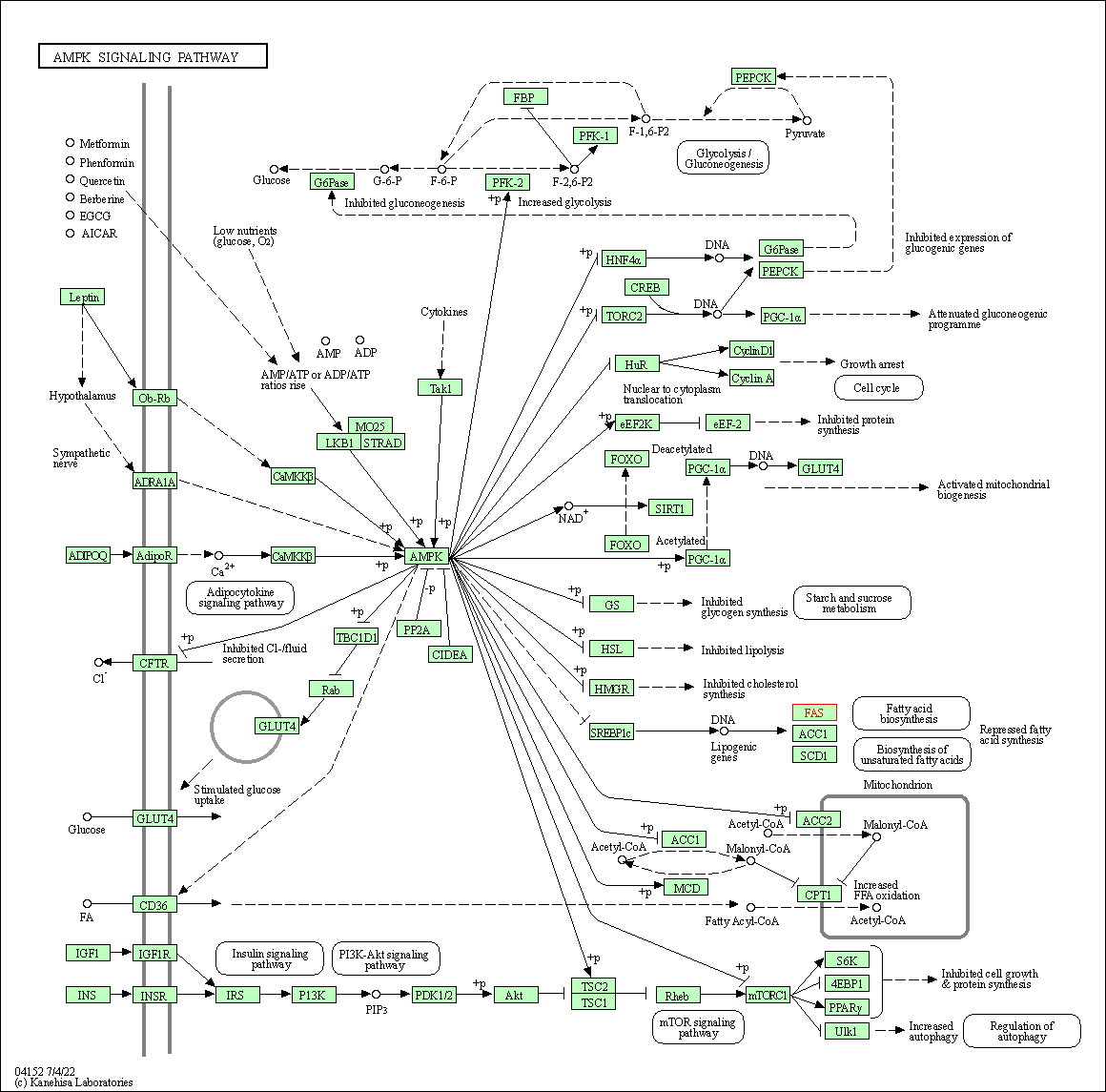
| AMPK signaling pathway | hsa04152 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
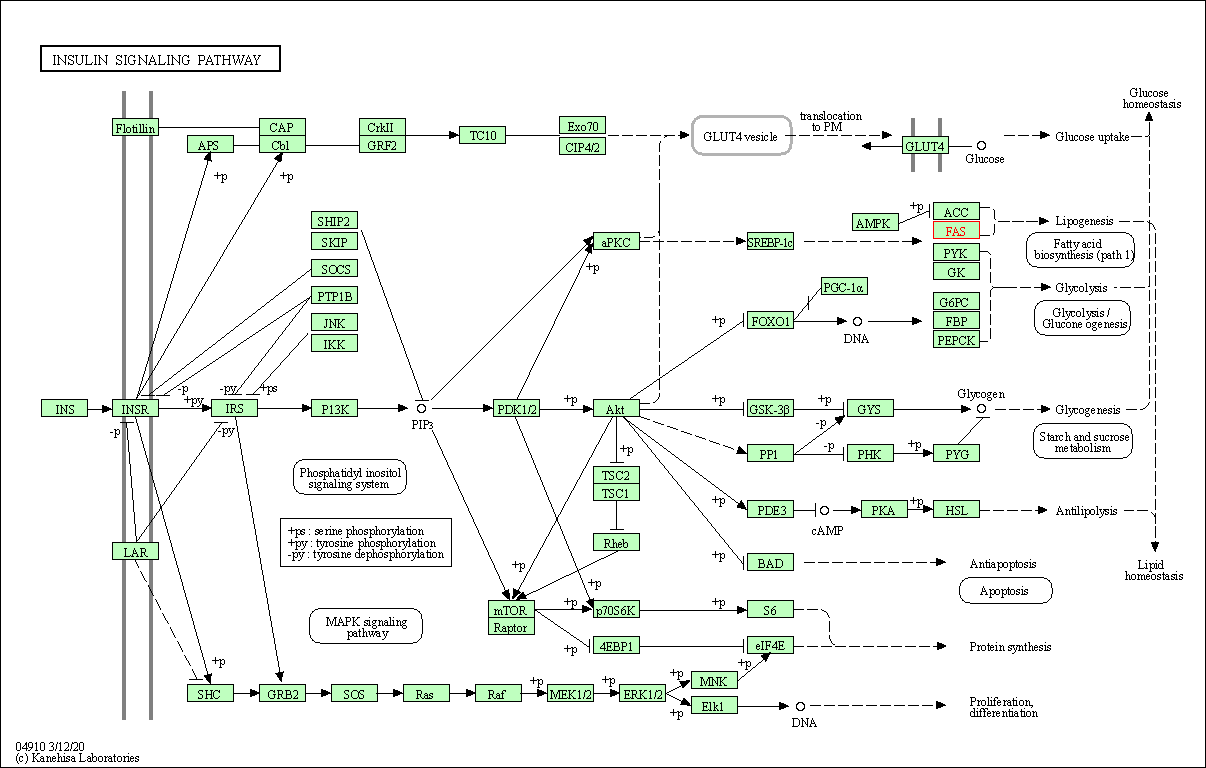
| Insulin signaling pathway | hsa04910 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 11 | Degree centrality | 1.18E-03 | Betweenness centrality | 7.94E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.99E-01 | Radiality | 1.34E+01 | Clustering coefficient | 1.45E-01 |
| Neighborhood connectivity | 9.82E+00 | Topological coefficient | 1.35E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Inhibition of fatty acid biosynthesis prevents adipocyte lipotoxicity on human osteoblasts in vitro. J Cell Mol Med. 2010 Apr;14(4):982-91. | |||||
| REF 2 | Novel agents in the management of Mycobacterium tuberculosis disease. Curr Med Chem. 2007;14(18):2000-8. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7002). | |||||
| REF 4 | The green tea polyphenol (2)-epigallocatechin-3-gallate (EGCG) is not a beta-secretase inhibitor. Bioorg Med Chem Lett. 2012 Feb 1;22(3):1408-14. | |||||
| REF 5 | Epigallocatechin gallate modulates CYP450 isoforms in the female Swiss-Webster mouse. Toxicol Sci. 2003 Dec;76(2):262-70. | |||||
| REF 6 | Prolyl endopeptidase inhibitors from green tea. Arch Pharm Res. 2001 Aug;24(4):292-6. | |||||
| REF 7 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 8 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 9 | ClinicalTrials.gov (NCT03938246) Study of TVB 2640 in Subjects With Non-Alcoholic Steatohepatitis (NASH). U.S. National Institutes of Health. | |||||
| REF 10 | Identification of a novel cell binding site of periostin involved in tumour growth. Eur J Cancer. 2011 Sep;47(14):2221-9. | |||||
| REF 11 | Proliferation inhibition and apoptosis induction of K562 cells by fatty acid synthase inhibitor--cerulenin. Zhonghua Xue Ye Xue Za Zhi. 2000 May;21(5):244-6. | |||||
| REF 12 | The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev. 1976 Sep;40(3):681-97. | |||||
| REF 13 | Inhibition of Plasmodium falciparum fatty acid biosynthesis: evaluation of FabG, FabZ, and FabI as drug targets for flavonoids. J Med Chem. 2006 Jun 1;49(11):3345-53. | |||||
| REF 14 | Imidazopyridine-Based Fatty Acid Synthase Inhibitors That Show Anti-HCV Activity and in Vivo Target Modulation. ACS Med Chem Lett. 2013 January 10; 4(1): 113-117. | |||||
| REF 15 | Fatty Acid Synthase Inhibitor TVB-2640 Reduces Hepatic de Novo Lipogenesis in Males With Metabolic Abnormalities. Hepatology. 2020 Jul;72(1):103-118. | |||||
| REF 16 | Novel antifungal agents, targets or therapeutic strategies for the treatment of invasive fungal diseases: a review of the literature (2005-2009). Rev Iberoam Micol. 2009 Mar 31;26(1):15-22. | |||||
| REF 17 | 2-Hexadecynoic acid inhibits plasmodial FAS-II enzymes and arrests erythrocytic and liver stage Plasmodium infections. Bioorg Med Chem. 2010 Nov 1;18(21):7475-85. | |||||
| REF 18 | 3-Aryl-4-hydroxyquinolin-2(1H)-one derivatives as type I fatty acid synthase inhibitors. Bioorg Med Chem Lett. 2006 Sep 1;16(17):4620-3. | |||||
| REF 19 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2608). | |||||
| REF 20 | C75 increases peripheral energy utilization and fatty acid oxidation in diet-induced obesity. Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9498-502. | |||||
| REF 21 | A human fatty acid synthase inhibitor binds beta-ketoacyl reductase in the keto-substrate site. Nat Chem Biol. 2014 Sep;10(9):774-9. | |||||
| REF 22 | Discovery and optimization of novel piperazines as potent inhibitors of fatty acid synthase (FASN). Bioorg Med Chem Lett. 2019 Apr 15;29(8):1001-1006. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

