Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T16538
|
|||||
| Target Name |
EGFR T790M mutant (EGFR T790M)
|
|||||
| Synonyms |
Receptor tyrosine-protein kinase erbB-1 T790M mutant; Proto-oncogene c-ErbB-1 T790M mutant; HER1 T790M mutant; Epidermal growth factor receptor T790M mutant; ERBB1 T790M mutant; ERBB T790M mutant
Click to Show/Hide
|
|||||
| Gene Name |
EGFR
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Lung cancer [ICD-11: 2C25] | |||||
| Function |
Receptor tyrosine kinase binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses. Known ligands include EGF, TGFA/TGF-alpha, AREG, epigen/EPGN, BTC/betacellulin, epiregulin/EREG and HBEGF/heparin-binding EGF. Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits adapter proteins like GRB2 which in turn activates complex downstream signaling cascades. Activates at least 4 major downstream signaling cascades including the RAS-RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC and STATs modules. May also activate the NF-kappa-B signaling cascade. Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling. Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/beta-catenin. Plays a role in enhancing learning and memory performance.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.10.1
|
|||||
| Sequence |
MRPSGTAGAALLALLAALCPASRALEEKKVCQGTSNKLTQLGTFEDHFLSLQRMFNNCEV
VLGNLEITYVQRNYDLSFLKTIQEVAGYVLIALNTVERIPLENLQIIRGNMYYENSYALA VLSNYDANKTGLKELPMRNLQEILHGAVRFSNNPALCNVESIQWRDIVSSDFLSNMSMDF QNHLGSCQKCDPSCPNGSCWGAGEENCQKLTKIICAQQCSGRCRGKSPSDCCHNQCAAGC TGPRESDCLVCRKFRDEATCKDTCPPLMLYNPTTYQMDVNPEGKYSFGATCVKKCPRNYV VTDHGSCVRACGADSYEMEEDGVRKCKKCEGPCRKVCNGIGIGEFKDSLSINATNIKHFK NCTSISGDLHILPVAFRGDSFTHTPPLDPQELDILKTVKEITGFLLIQAWPENRTDLHAF ENLEIIRGRTKQHGQFSLAVVSLNITSLGLRSLKEISDGDVIISGNKNLCYANTINWKKL FGTSGQKTKIISNRGENSCKATGQVCHALCSPEGCWGPEPRDCVSCRNVSRGRECVDKCN LLEGEPREFVENSECIQCHPECLPQAMNITCTGRGPDNCIQCAHYIDGPHCVKTCPAGVM GENNTLVWKYADAGHVCHLCHPNCTYGCTGPGLEGCPTNGPKIPSIATGMVGALLLLLVV ALGIGLFMRRRHIVRKRTLRRLLQERELVEPLTPSGEAPNQALLRILKETEFKKIKVLGS GAFGTVYKGLWIPEGEKVKIPVAIKELREATSPKANKEILDEAYVMASVDNPHVCRLLGI CLTSTVQLITQLMPFGCLLDYVREHKDNIGSQYLLNWCVQIAKGMNYLEDRRLVHRDLAA RNVLVKTPQHVKITDFGLAKLLGAEEKEYHAEGGKVPIKWMALESILHRIYTHQSDVWSY GVTVWELMTFGSKPYDGIPASEISSILEKGERLPQPPICTIDVYMIMVKCWMIDADSRPK FRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDMDDVVDADEYLIPQ QGFFSSPSTSRTPLLSSLSATSNNSTVACIDRNGLQSCPIKEDSFLQRYSSDPTGALTED SIDDTFLPVPEYINQSVPKRPAGSVQNPVYHNQPLNPAPSRDPHYQDPHSTAVGNPEYLN TVQPTCVNSTFDSPAHWAQKGSHQISLDNPDYQQDFFPKEAKPNGIFKGSTAENAEYLRV APQSSEFIGA Click to Show/Hide
|
|||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Osimertinib | Drug Info | Approved | Non-small-cell lung cancer | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 2 Inhibitor drugs | + | ||||
| 1 | Osimertinib | Drug Info | [3] | |||
| 2 | PMID27774824-Compound-Figure9Example2down | Drug Info | [1] | |||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Gefitinib | Ligand Info | |||||
| Structure Description | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with gefitinib | PDB:3UG2 | ||||
| Method | X-ray diffraction | Resolution | 2.50 Å | Mutation | Yes | [4] |
| PDB Sequence |
EAPNQALLRI
706 LKETEFKKIK716 VLSSGTVYKG729 LWIPEGEKVK739 IPVAIKESPK754 ANKEILDEAY 764 VMASVDNPHV774 CRLLGICLTS784 TVQLIMQLMP794 FGCLLDYVRE804 HKDNIGSQYL 814 LNWCVQIAKG824 MNYLEDRRLV834 HRDLAARNVL844 VKTPQHVKIT854 DFGLAKLLGA 864 EEKEYHAEGG874 KVPIKWMALE884 SILHRIYTHQ894 SDVWSYGVTV904 WELMTFGSKP 914 YDGIPASEIS924 SILEKGERLP934 QPPICTIDVY944 MIMVKCWMID954 ADSRPKFREL 964 IIEFSKMARD974 PQRYLVIQGD984 ERMHLEDMDD1009 VVDADEYLI
|
|||||
|
|
||||||
| Ligand Name: Ibrutinib | Ligand Info | |||||
| Structure Description | Crystal structure of EGFR 696-1022 T790M in complex with Ibrutinib | PDB:5YU9 | ||||
| Method | X-ray diffraction | Resolution | 1.95 Å | Mutation | Yes | [5] |
| PDB Sequence |
NQALLRILKE
709 TEFKKIKVLG719 SGAFGTVYKG729 LWIPEGEKVK739 IPVAIKELRE749 KANKEILDEA 763 YVMASVDNPH773 VCRLLGICLT783 STVQLIMQLM793 PFGCLLDYVR803 EHKDNIGSQY 813 LLNWCVQIAK823 GMNYLEDRRL833 VHRDLAARNV843 LVKTPQHVKI853 TDFGLAKLLK 875 VPIKWMALES885 ILHRIYTHQS895 DVWSYGVTVW905 ELMTFGSKPY915 DGIPASEISS 925 ILEKGERLPQ935 PPICTIDVYM945 IMVKCWMIDA955 DSRPKFRELI965 IEFSKMARDP 975 QRYLVIQGDE985 RMHLPSPTDS995 NFYRALMDEE1005 DMDDVVDADE1015 YL |
|||||
|
|
LEU718
3.651
GLY719
3.593
SER720
4.926
VAL726
3.800
ALA743
3.201
ILE744
4.874
LYS745
3.834
MET766
3.484
CYS775
4.182
ARG776
4.780
LEU777
4.330
LEU788
3.554
MET790
3.296
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Pathway Affiliation
|
|
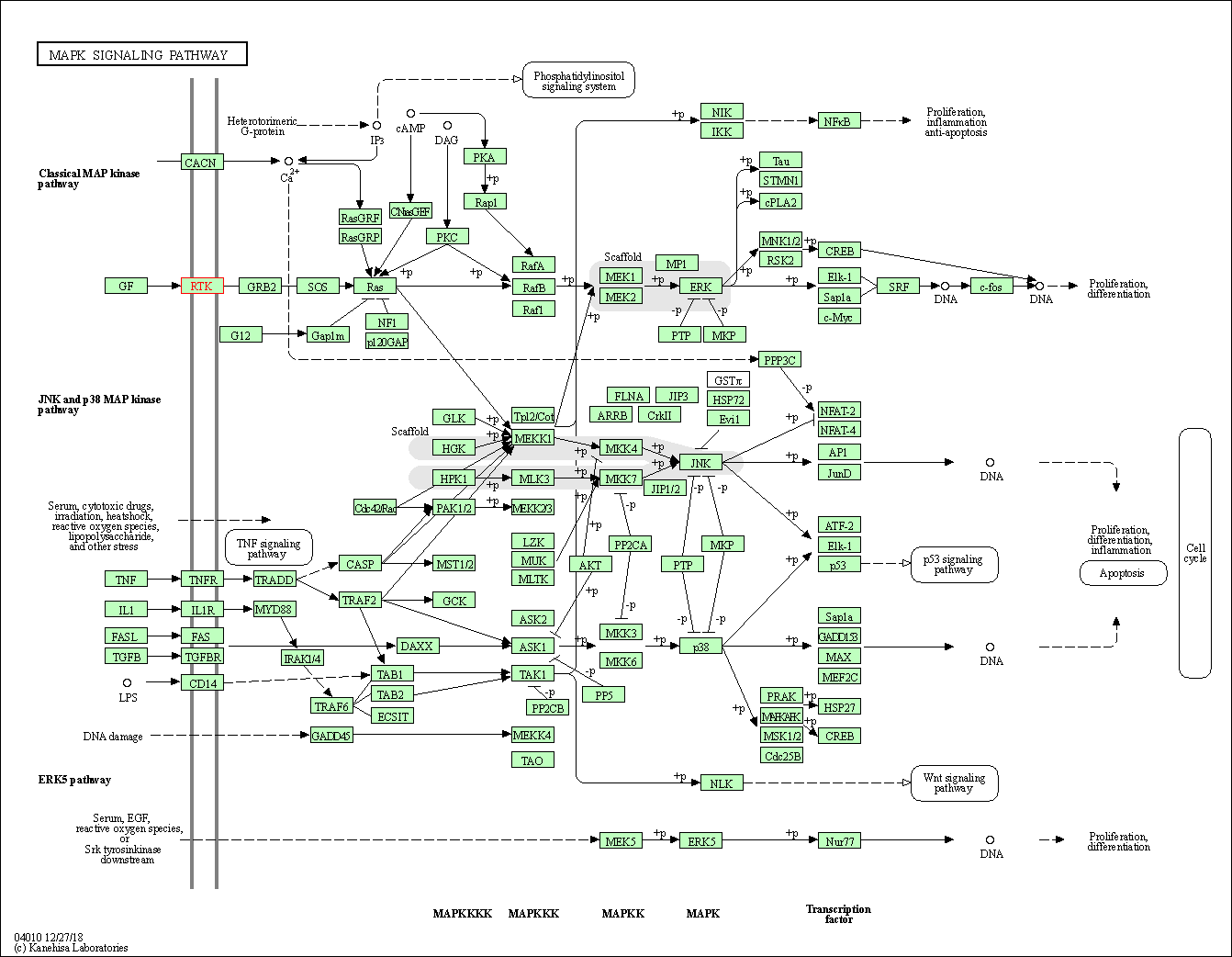
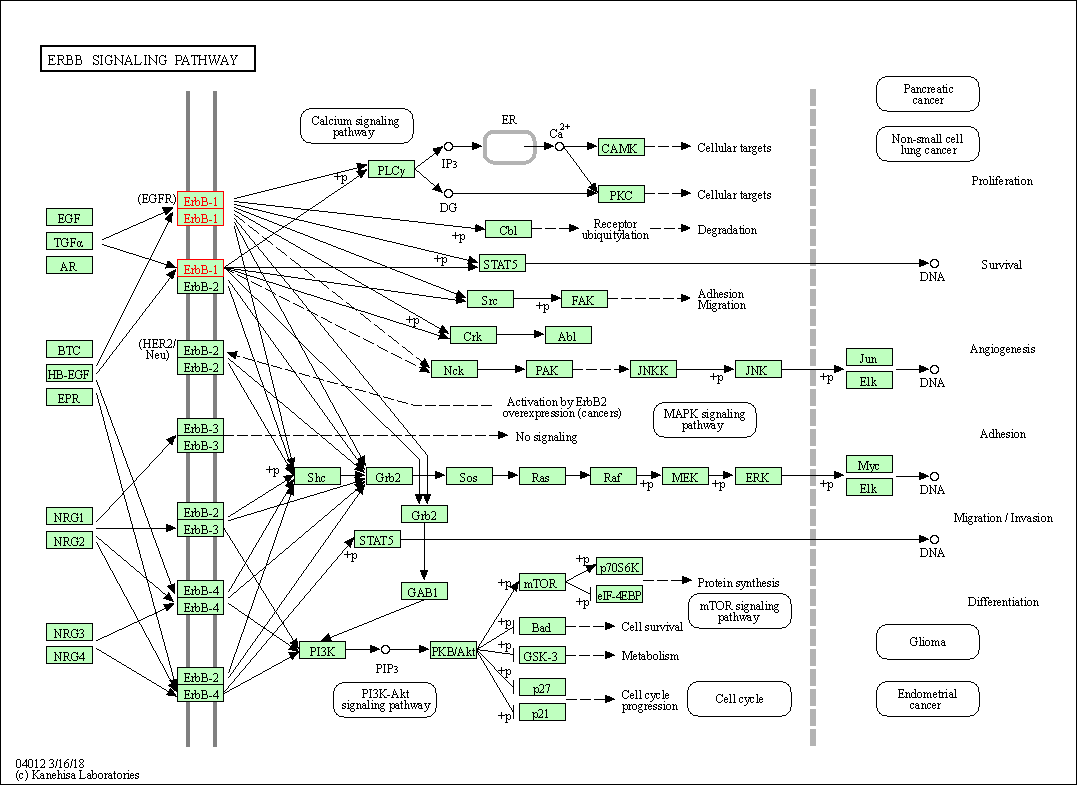
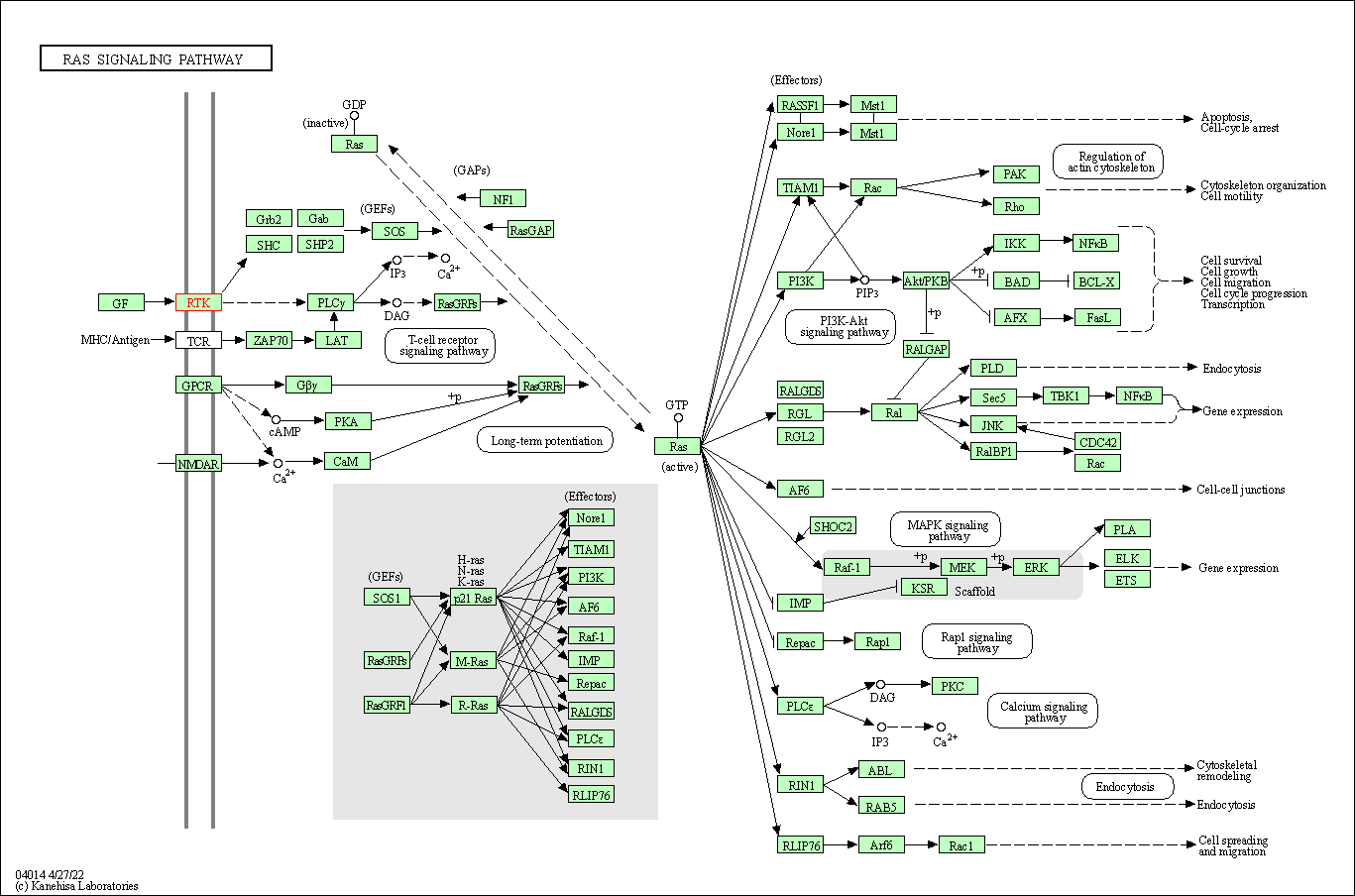
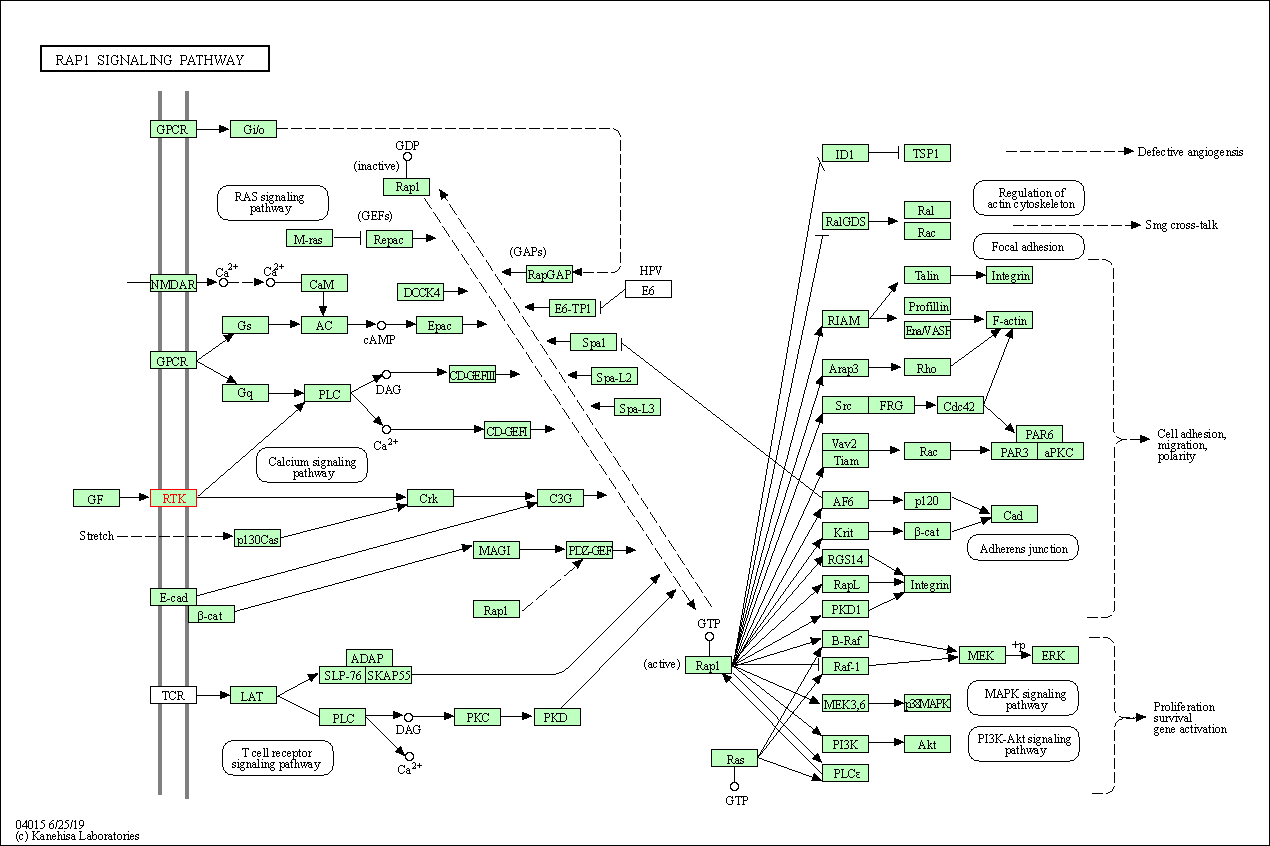
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| ErbB signaling pathway | hsa04012 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Calcium signaling pathway | hsa04020 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Endocytosis | hsa04144 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Adherens junction | hsa04520 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Gap junction | hsa04540 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| JAK-STAT signaling pathway | hsa04630 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Parathyroid hormone synthesis, secretion and action | hsa04928 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Inhibitors of JAK-family kinases: an update on the patent literature 2013-2015, part 1.Expert Opin Ther Pat. 2017 Feb;27(2):127-143. | |||||
| REF 2 | The ChEMBL database in 2017. Nucleic Acids Res. 2017 Jan 4;45(D1):D945-D954. | |||||
| REF 3 | AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014 Sep;4(9):1046-61. | |||||
| REF 4 | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene. 2013 Jan 3;32(1):27-38. | |||||
| REF 5 | Ibrutinib targets mutant-EGFR kinase with a distinct binding conformation. Oncotarget. 2016 Oct 25;7(43):69760-69769. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

