Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T20953
(Former ID: TTDI03066)
|
|||||
| Target Name |
BUB1 mitotic checkpoint serine/threonine kinase (BUB1)
|
|||||
| Synonyms |
hBUB1; Mitotic checkpoint serine/threonine-protein kinase BUB1; BUB1L; BUB1A
Click to Show/Hide
|
|||||
| Gene Name |
BUB1
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Function |
Has a key role in the assembly of checkpoint proteins at the kinetochore, being required for the subsequent localization of CENPF, BUB1B, CENPE and MAD2L1. Required for the kinetochore localization of PLK1. Required for centromeric enrichment of AUKRB in prometaphase. Plays an important role in defining SGO1 localization and thereby affects sister chromatid cohesion. Acts as a substrate for anaphase-promoting complex or cyclosome (APC/C) in complex with its activator CDH1 (APC/C-Cdh1). Necessary for ensuring proper chromosome segregation and binding to BUB3 is essential for this function. Can regulate chromosome segregation in a kinetochore-independent manner. Can phosphorylate BUB3. The BUB1-BUB3 complex plays a role in the inhibition of APC/C when spindle-assembly checkpoint is activated and inhibits the ubiquitin ligase activity of APC/C by phosphorylating its activator CDC20. This complex can also phosphorylate MAD1L1. Kinase activity is essential for inhibition of APC/CCDC20 and for chromosome alignment but does not play a major role in the spindle-assembly checkpoint activity. Mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. Serine/threonine-protein kinase that performs 2 crucial functions during mitosis: it is essential for spindle-assembly checkpoint signaling and for correct chromosome alignment.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MDTPENVLQMLEAHMQSYKGNDPLGEWERYIQWVEENFPENKEYLITLLEHLMKEFLDKK
KYHNDPRFISYCLKFAEYNSDLHQFFEFLYNHGIGTLSSPLYIAWAGHLEAQGELQHASA VLQRGIQNQAEPREFLQQQYRLFQTRLTETHLPAQARTSEPLHNVQVLNQMITSKSNPGN NMACISKNQGSELSGVISSACDKESNMERRVITISKSEYSVHSSLASKVDVEQVVMYCKE KLIRGESEFSFEELRAQKYNQRRKHEQWVNEDRHYMKRKEANAFEEQLLKQKMDELHKKL HQVVETSHEDLPASQERSEVNPARMGPSVGSQQELRAPCLPVTYQQTPVNMEKNPREAPP VVPPLANAISAALVSPATSQSIAPPVPLKAQTVTDSMFAVASKDAGCVNKSTHEFKPQSG AEIKEGCETHKVANTSSFHTTPNTSLGMVQATPSKVQPSPTVHTKEALGFIMNMFQAPTL PDISDDKDEWQSLDQNEDAFEAQFQKNVRSSGAWGVNKIISSLSSAFHVFEDGNKENYGL PQPKNKPTGARTFGERSVSRLPSKPKEEVPHAEEFLDDSTVWGIRCNKTLAPSPKSPGDF TSAAQLASTPFHKLPVESVHILEDKENVVAKQCTQATLDSCEENMVVPSRDGKFSPIQEK SPKQALSSHMYSASLLRLSQPAAGGVLTCEAELGVEACRLTDTDAAIAEDPPDAIAGLQA EWMQMSSLGTVDAPNFIVGNPWDDKLIFKLLSGLSKPVSSYPNTFEWQCKLPAIKPKTEF QLGSKLVYVHHLLGEGAFAQVYEATQGDLNDAKNKQKFVLKVQKPANPWEFYIGTQLMER LKPSMQHMFMKFYSAHLFQNGSVLVGELYSYGTLLNAINLYKNTPEKVMPQGLVISFAMR MLYMIEQVHDCEIIHGDIKPDNFILGNGFLEQDDEDDLSAGLALIDLGQSIDMKLFPKGT IFTAKCETSGFQCVEMLSNKPWNYQIDYFGVAATVYCMLFGTYMKVKNEGGECKPEGLFR RLPHLDMWNEFFHVMLNIPDCHHLPSLDLLRQKLKKVFQQHYTNKIRALRNRLIVLLLEC KRSRK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T75I26 | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: L-serine-O-phosphate | Ligand Info | |||||
| Structure Description | Crystal structure of the human Bub1 kinase domain in complex with BAY 1816032 | PDB:6F7B | ||||
| Method | X-ray diffraction | Resolution | 2.00 Å | Mutation | No | [3] |
| PDB Sequence |
APNFIVGNPW
742 DDKLIFKLLS752 GLSKPVSSYP762 NTFEWQCKLP772 AIKPKTEFQL782 GSKLVYVHHL 792 LGEGAFAQVY802 EATQKNKQKF818 VLKVQKPANP828 WEFYIGTQLM838 ERLKPSMQHM 848 FMKFYSAHLF858 QNGSVLVGEL868 YSYGTLLNAI878 NLYKNTPEKV888 MPQGLVISFA 898 MRMLYMIEQV908 HDCEIIHGDI918 KPDNFILGNG928 FLEQDDEDDL938 SAGLALIDLG 948 QSIDMKLFPK958 GTIFTAKCET968 GFQCVEMLSN979 KPWNYQIDYF989 GVAATVYCML 999 FGTYMKVKNE1009 ECKPEGLFRR1021 LPHLDMWNEF1031 FHVMLNIPDC1041 HHLPSLDLLR 1051 QKLKKVFQQH1061 YTNKIRALRN1071 RLIVLLLECK1081 RS
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: adenosine diphosphate | Ligand Info | |||||
| Structure Description | Structure of Bub1 kinase domain | PDB:4QPM | ||||
| Method | X-ray diffraction | Resolution | 2.20 Å | Mutation | No | [4] |
| PDB Sequence |
PNPWDDKLIF
748 KLLSGLSKPV758 SSYPNTFEWQ768 CKLPAIKPKT778 EFQLGSKLVY788 VHHLLGEGAF 798 AQVYEATQKQ816 KFVLKVQKPA826 NPWEFYIGTQ836 LMERLKPSMQ846 HMFMKFYSAH 856 LFQNGSVLVG866 ELYSYGTLLN876 AINLYKNTPE886 KVMPQGLVIS896 FAMRMLYMIE 906 QVHDCEIIHG916 DIKPDNFILG926 NGFLESAGLA943 LIDLGQSIDM953 KLFPKGTIFT 963 AKCETGFQCV974 EMLSNKPWNY984 QIDYFGVAAT994 VYCMLFGTYM1004 KVKNEGGECK 1014 PEGLFRRLPH1024 LDMWNEFFHV1034 MLNIPDCHHL1044 PSLDLLRQKL1054 KKVFQQHYTN 1064 KIRALRNRLI1074 VLLLECKRS
|
|||||
|
|
LEU793
2.425
GLY794
2.616
GLU795
3.000
GLY796
2.235
ALA797
2.685
PHE798
2.075
ALA799
2.057
GLN800
4.341
VAL801
2.532
VAL819
3.002
LYS821
1.861
GLU830
4.470
MET850
2.717
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cell cycle | hsa04110 | Affiliated Target |
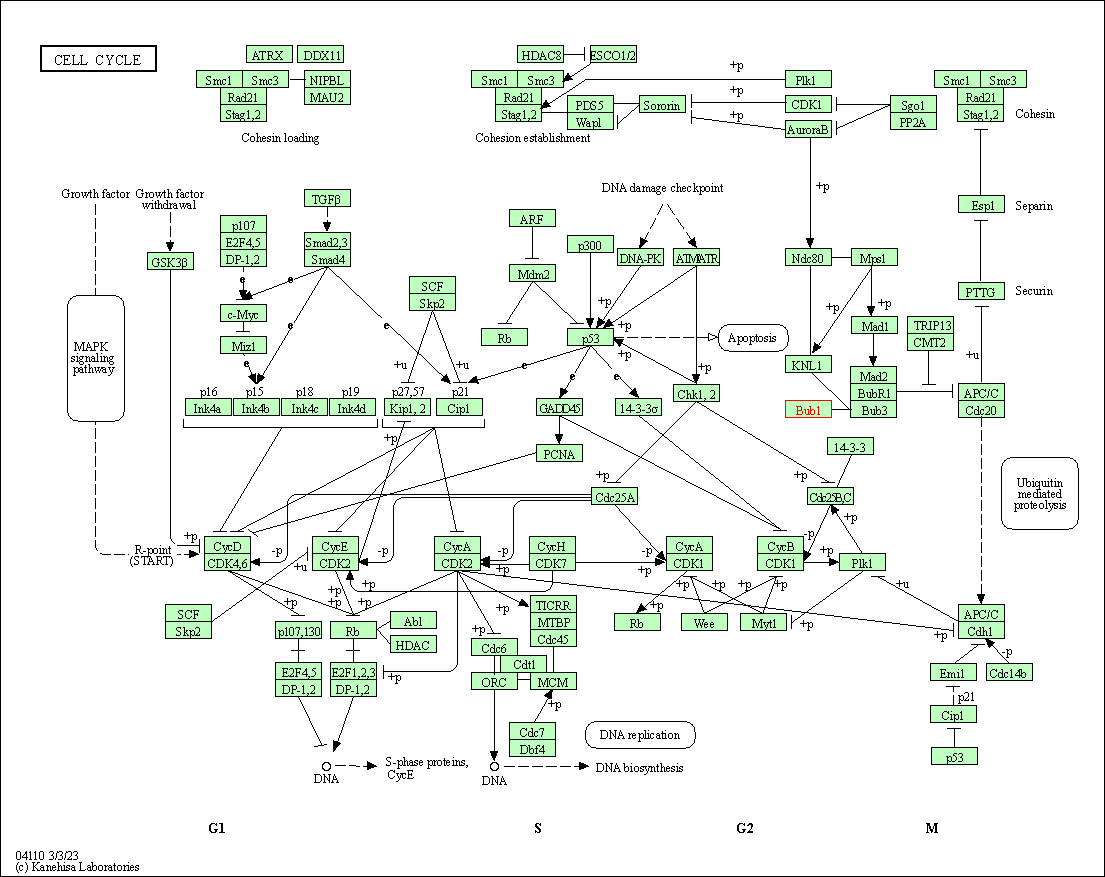
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |
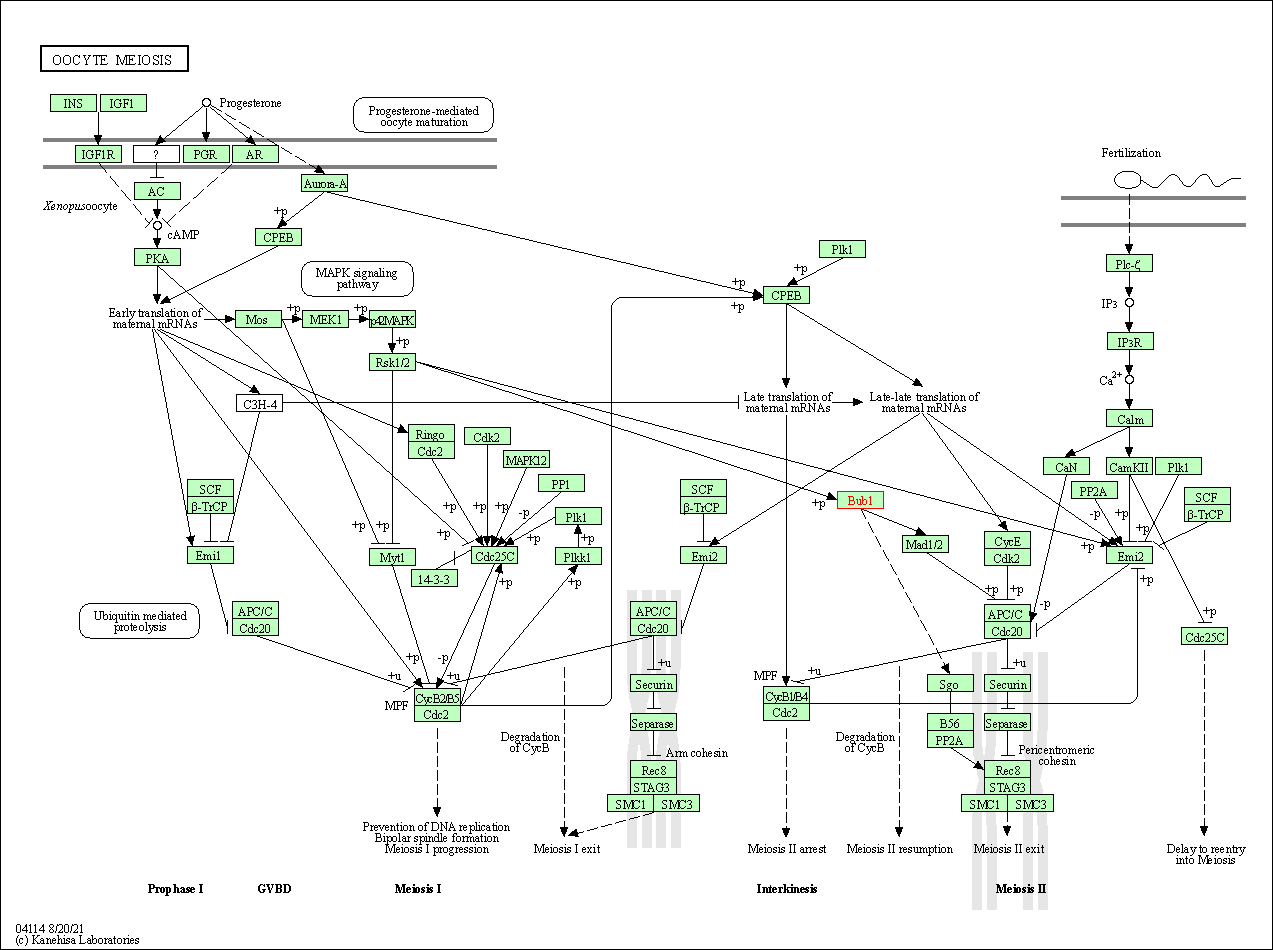
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |
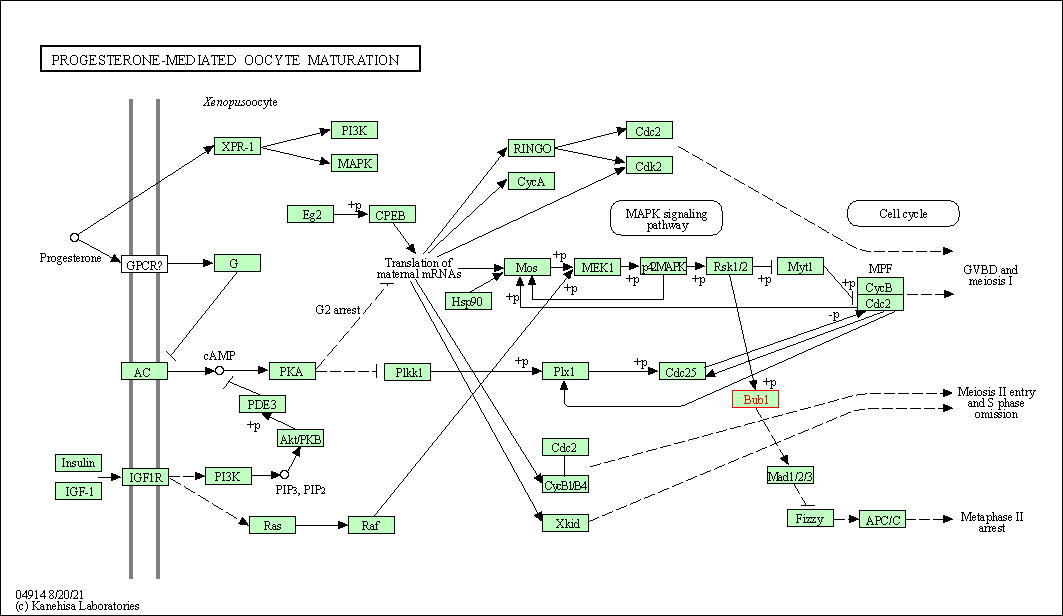
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 56 | Degree centrality | 6.02E-03 | Betweenness centrality | 6.65E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.23E-01 | Radiality | 1.39E+01 | Clustering coefficient | 3.79E-01 |
| Neighborhood connectivity | 3.64E+01 | Topological coefficient | 1.01E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Novel cycloalkenepyrazoles as inhibitors of bub1 kinase. ACS Med Chem Lett. 2014 Feb 12;5(4):280-1. | |||||
| REF 2 | Substituted benzylpyrazoles. US9765058. | |||||
| REF 3 | Inhibition of BUB1 Kinase by BAY 1816032 Sensitizes Tumor Cells toward Taxanes, ATR, and PARP Inhibitors In Vitro and In Vivo. Clin Cancer Res. 2019 Feb 15;25(4):1404-1414. | |||||
| REF 4 | Substrate-specific activation of the mitotic kinase Bub1 through intramolecular autophosphorylation and kinetochore targeting. Structure. 2014 Nov 4;22(11):1616-27. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

