Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T63609
(Former ID: TTDS00500)
|
|||||
| Target Name |
Adenylate cyclase type 1 (ADCY1)
|
|||||
| Synonyms |
Ca(2+)/calmodulin-activated adenylyl cyclase; Adenylyl cyclase 1; Adenylate cyclase type I; ATP pyrophosphate-lyase 1
Click to Show/Hide
|
|||||
| Gene Name |
ADCY1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Nutritional deficiency [ICD-11: 5B50-5B71] | |||||
| Function |
Mediates responses to increased cellular Ca(2+)/calmodulin levels. May be involved in regulatory processes in the central nervous system. May play a role in memory and learning. Plays a role in the regulation of the circadian rhythm of daytime contrast sensitivity probably by modulating the rhythmic synthesis of cyclic AMP in the retina. Catalyzes the formation of the signaling molecule cAMP in response to G-protein signaling.
Click to Show/Hide
|
|||||
| BioChemical Class |
Phosphorus-oxygen lyase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 4.6.1.1
|
|||||
| Sequence |
MAGAPRGGGGGGGGAGEPGGAERAAGTSRRRGLRACDEEFACPELEALFRGYTLRLEQAA
TLKALAVLSLLAGALALAELLGAPGPAPGLAKGSHPVHCVLFLALLVVTNVRSLQVPQLQ QVGQLALLFSLTFALLCCPFALGGPARGSAGAAGGPATAEQGVWQLLLVTFVSYALLPVR SLLAIGFGLVVAASHLLVTATLVPAKRPRLWRTLGANALLFVGVNMYGVFVRILTERSQR KAFLQARSCIEDRLRLEDENEKQERLLMSLLPRNVAMEMKEDFLKPPERIFHKIYIQRHD NVSILFADIVGFTGLASQCTAQELVKLLNELFGKFDELATENHCRRIKILGDCYYCVSGL TQPKTDHAHCCVEMGLDMIDTITSVAEATEVDLNMRVGLHTGRVLCGVLGLRKWQYDVWS NDVTLANVMEAAGLPGKVHITKTTLACLNGDYEVEPGYGHERNSFLKTHNIETFFIVPSH RRKIFPGLILSDIKPAKRMKFKTVCYLLVQLMHCRKMFKAEIPFSNVMTCEDDDKRRALR TASEKLRNRSSFSTNVVYTTPGTRVNRYISRLLEARQTELEMADLNFFTLKYKHVEREQK YHQLQDEYFTSAVVLTLILAALFGLVYLLIFPQSVVVLLLLVFCICFLVACVLYLHITRV QCFPGCLTIQIRTVLCIFIVVLIYSVAQGCVVGCLPWAWSSKPNSSLVVLSSGGQRTALP TLPCESTHHALLCCLVGTLPLAIFFRVSSLPKMILLSGLTTSYILVLELSGYTRTGGGAV SGRSYEPIVAILLFSCALALHARQVDIRLRLDYLWAAQAEEEREDMEKVKLDNRRILFNL LPAHVAQHFLMSNPRNMDLYYQSYSQVGVMFASIPNFNDFYIELDGNNMGVECLRLLNEI IADFDELMEKDFYKDIEKIKTIGSTYMAAVGLAPTSGTKAKKSISSHLSTLADFAIEMFD VLDEINYQSYNDFVLRVGINVGPVVAGVIGARRPQYDIWGNTVNVASRMDSTGVQGRIQV TEEVHRLLRRCPYHFVCRGKVSVKGKGEMLTYFLEGRTDGNGSQIRSLGLDRKMCPFGRA GLQGRRPPVCPMPGVSVRAGLPPHSPGQYLPSAAAGKEA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A01671 ; BADD_A04410 ; BADD_A05200 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Adenosine monophosphate | Drug Info | Approved | Malnutrition | [2], [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Binder | [+] 1 Binder drugs | + | ||||
| 1 | Adenosine monophosphate | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
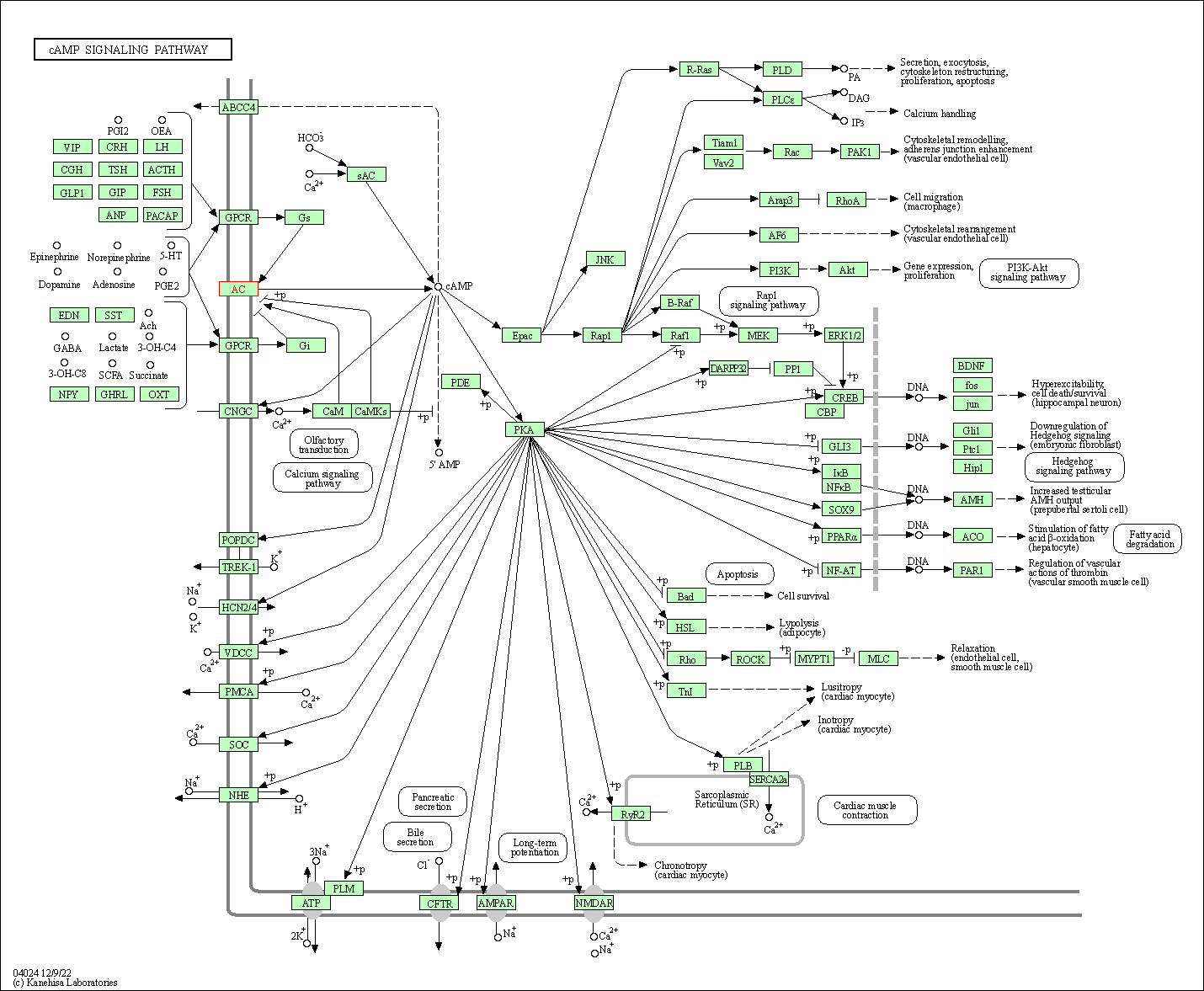
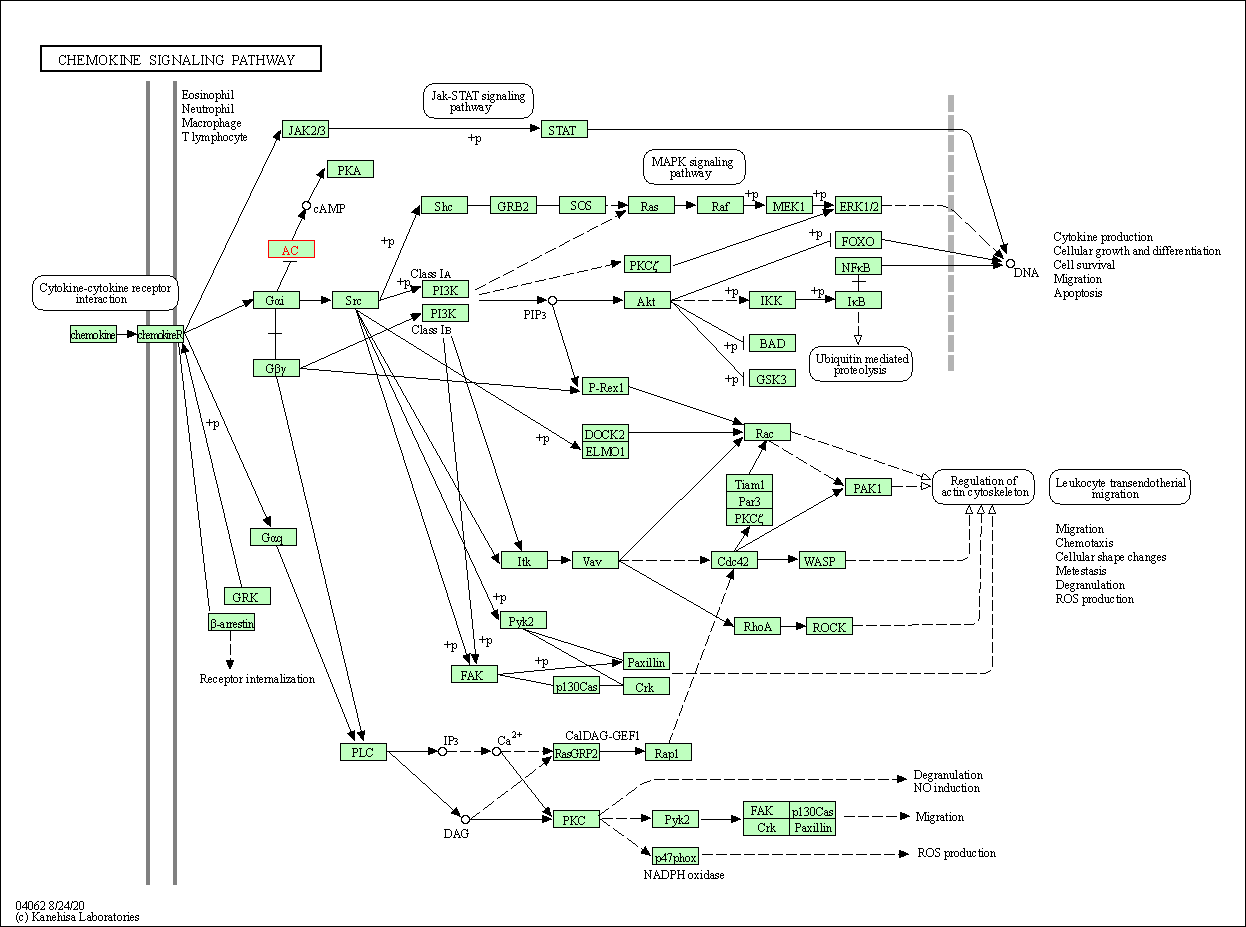
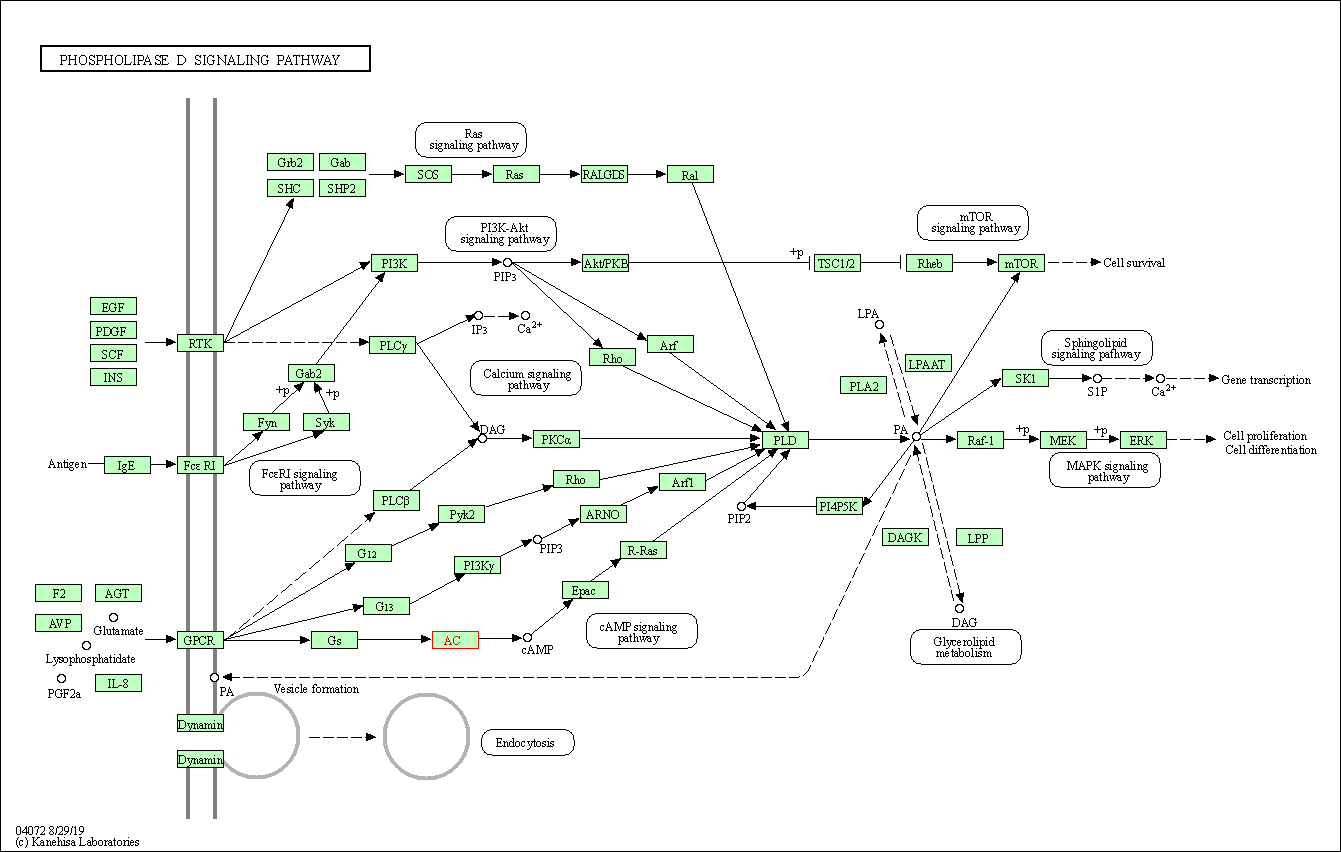

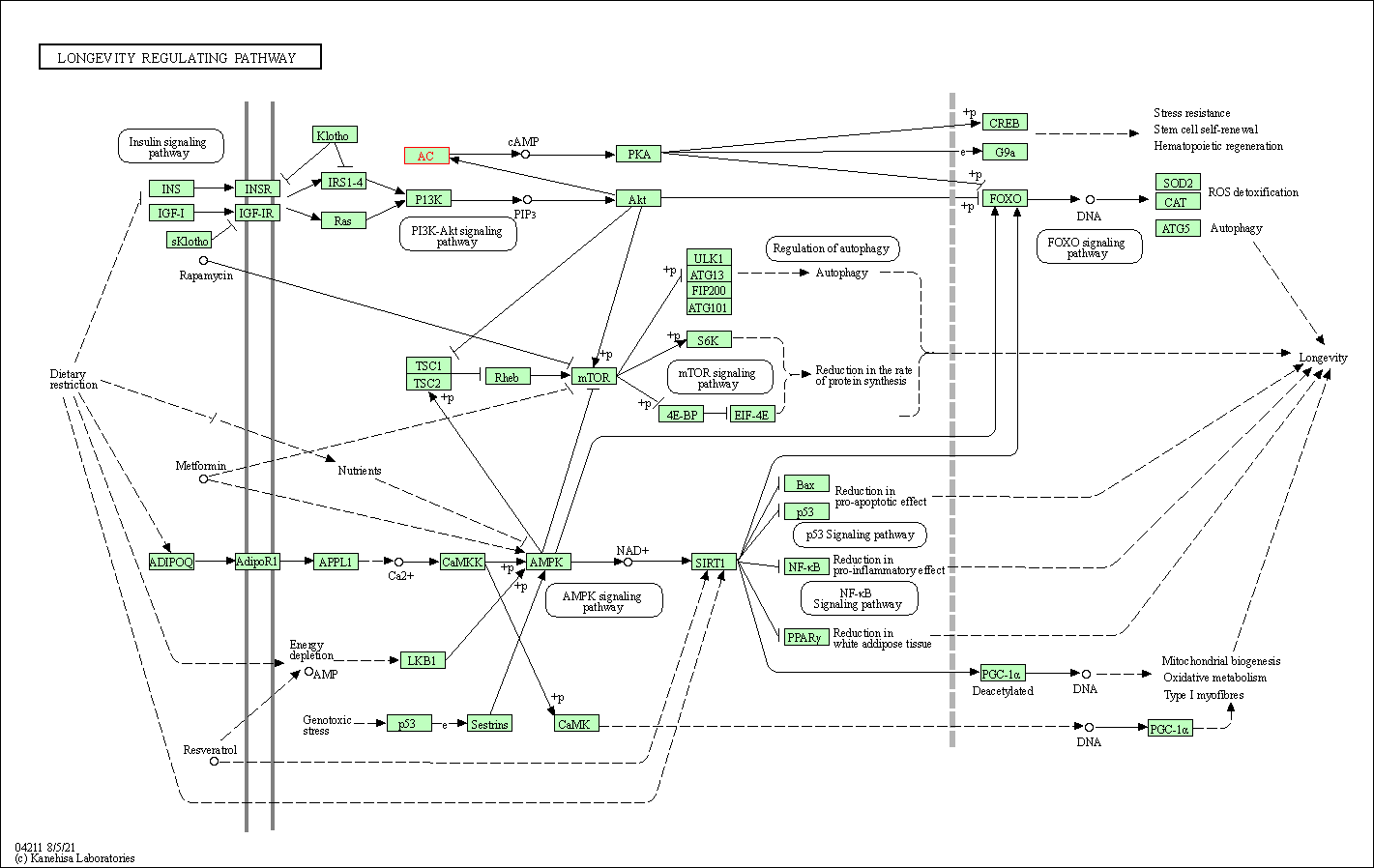




| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Purine metabolism | hsa00230 | Affiliated Target |

|
| Class: Metabolism => Nucleotide metabolism | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Calcium signaling pathway | hsa04020 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Chemokine signaling pathway | hsa04062 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
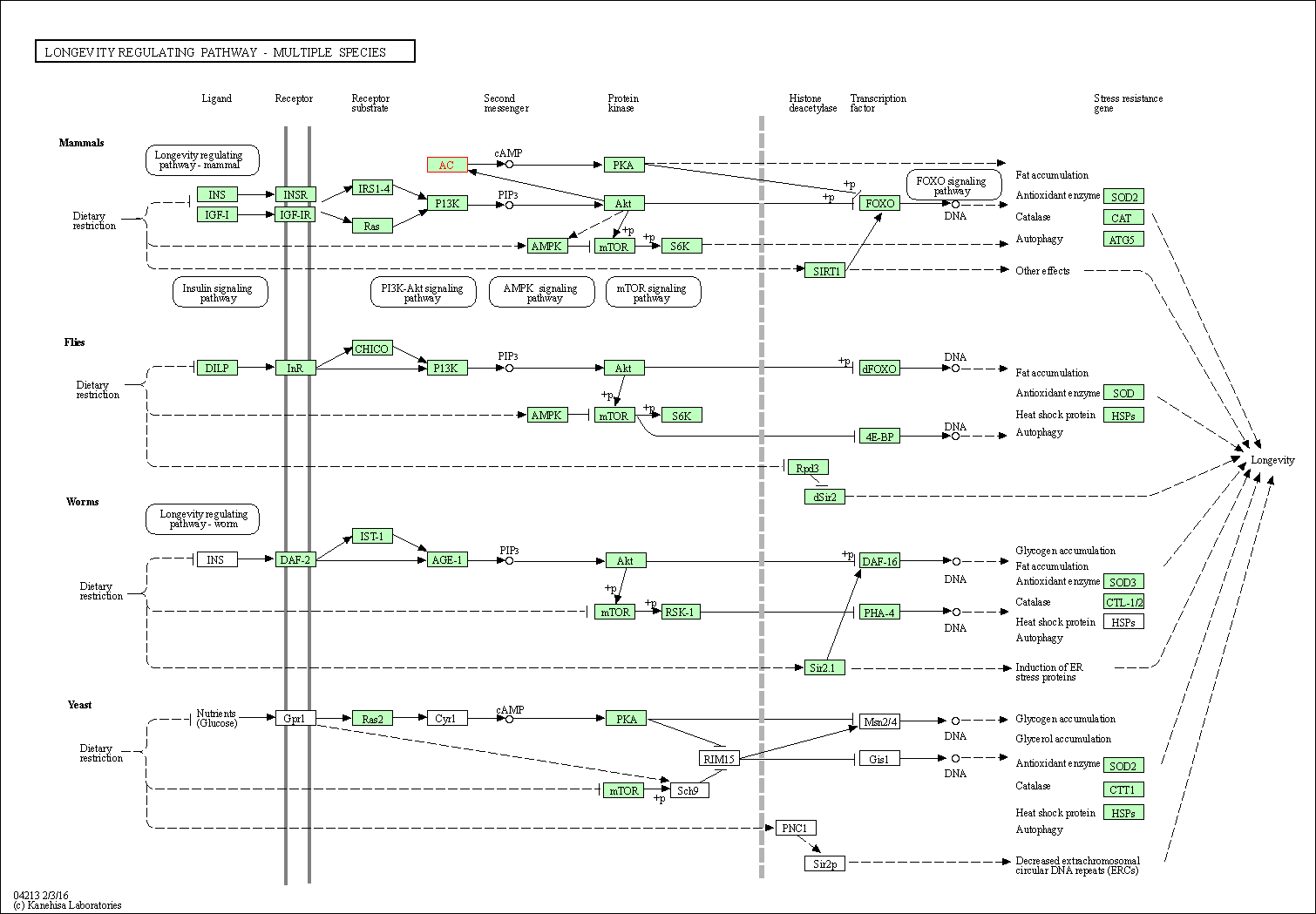
| Longevity regulating pathway | hsa04211 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Longevity regulating pathway - multiple species | hsa04213 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
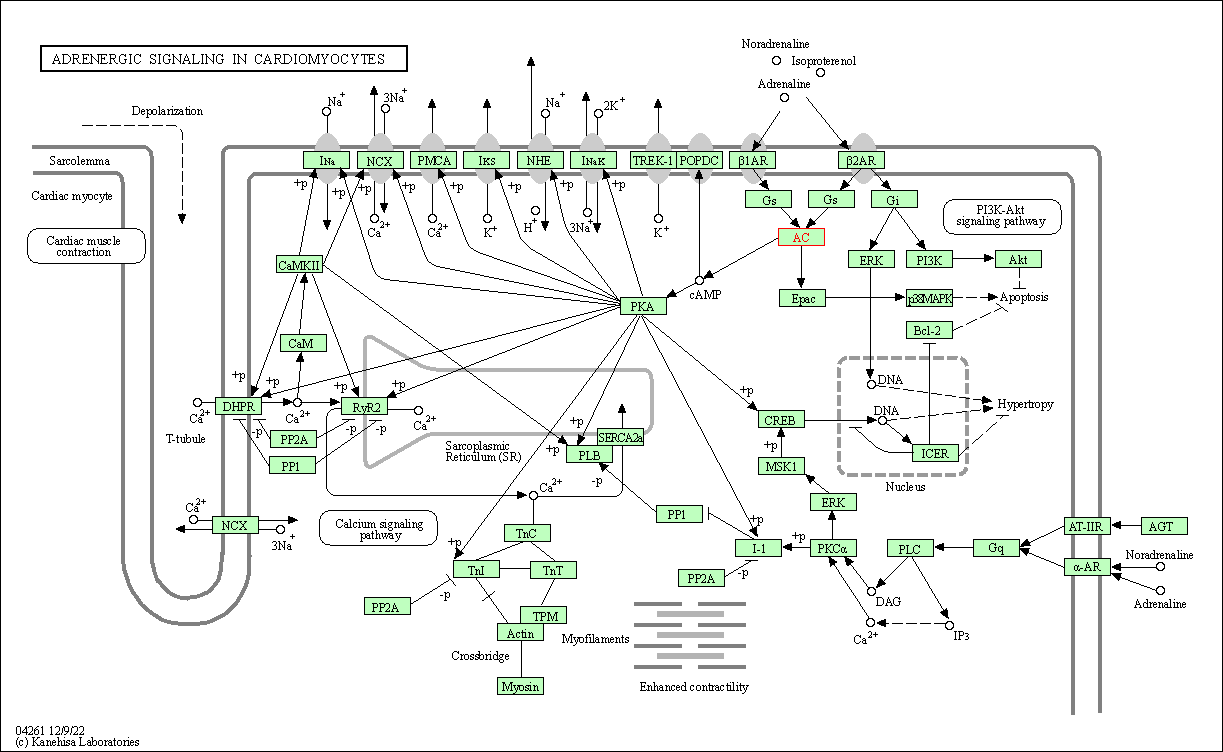
| Adrenergic signaling in cardiomyocytes | hsa04261 | Affiliated Target |

|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
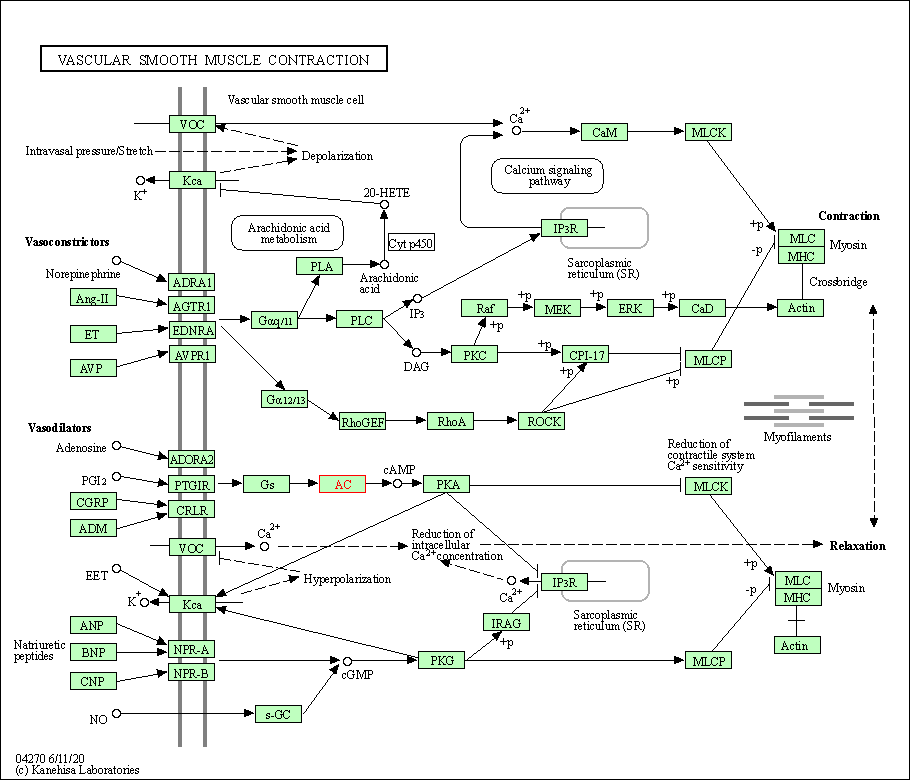
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |

|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
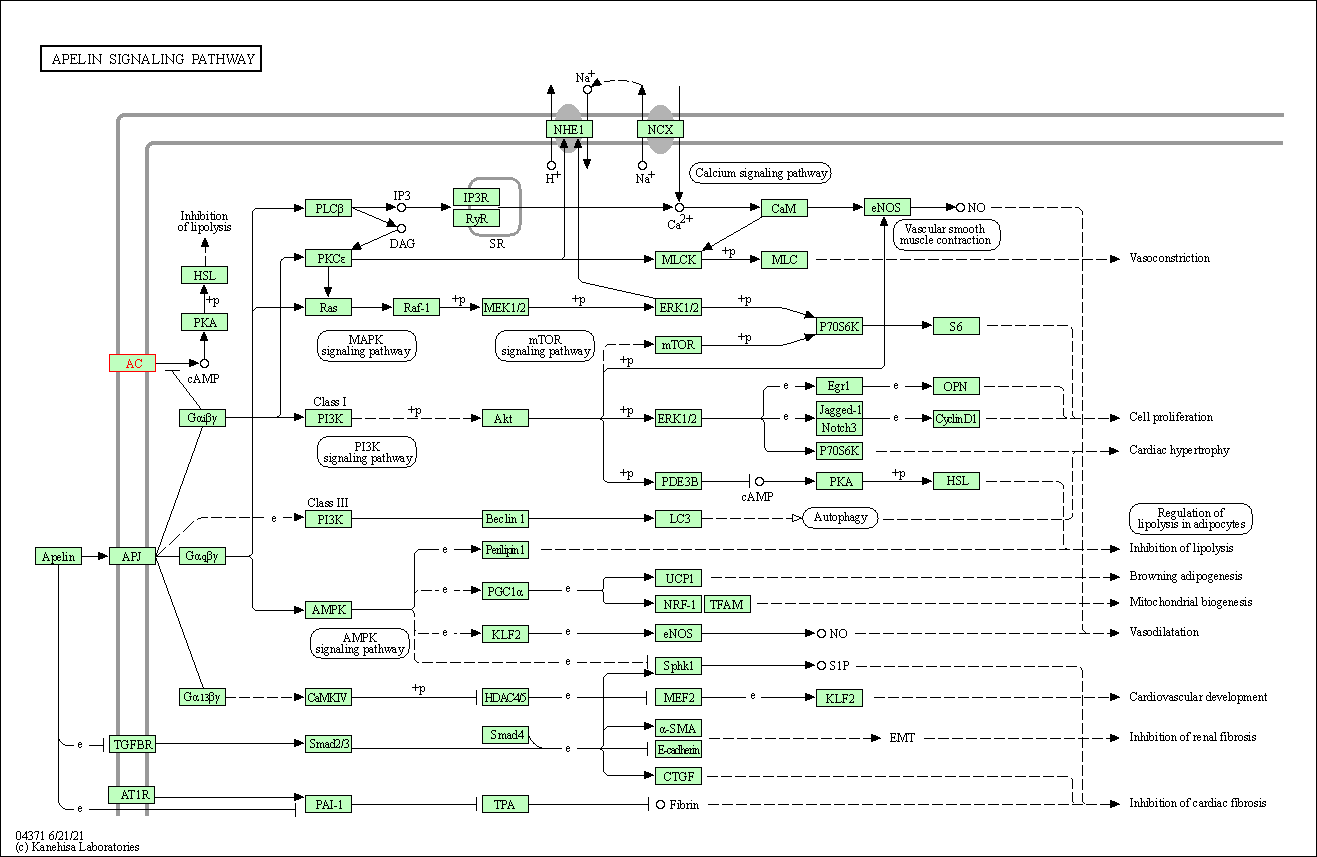
| Apelin signaling pathway | hsa04371 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
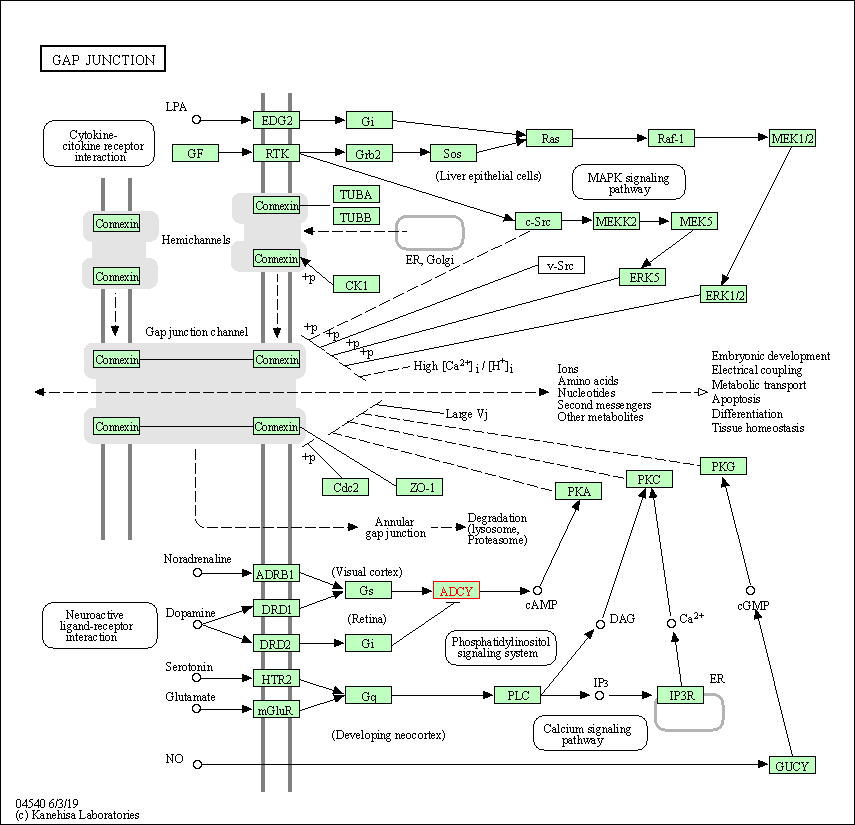
| Gap junction | hsa04540 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
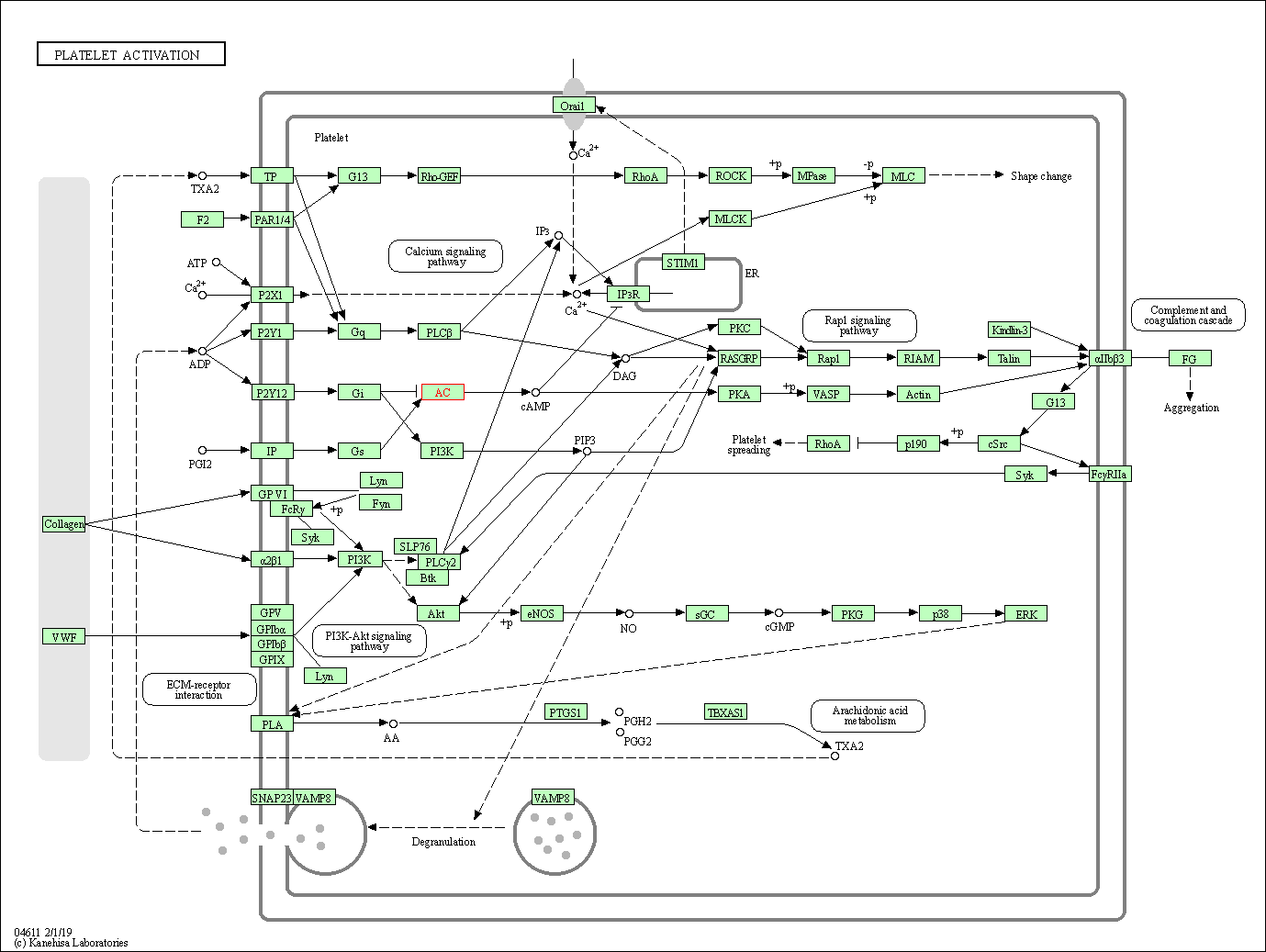
| Platelet activation | hsa04611 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
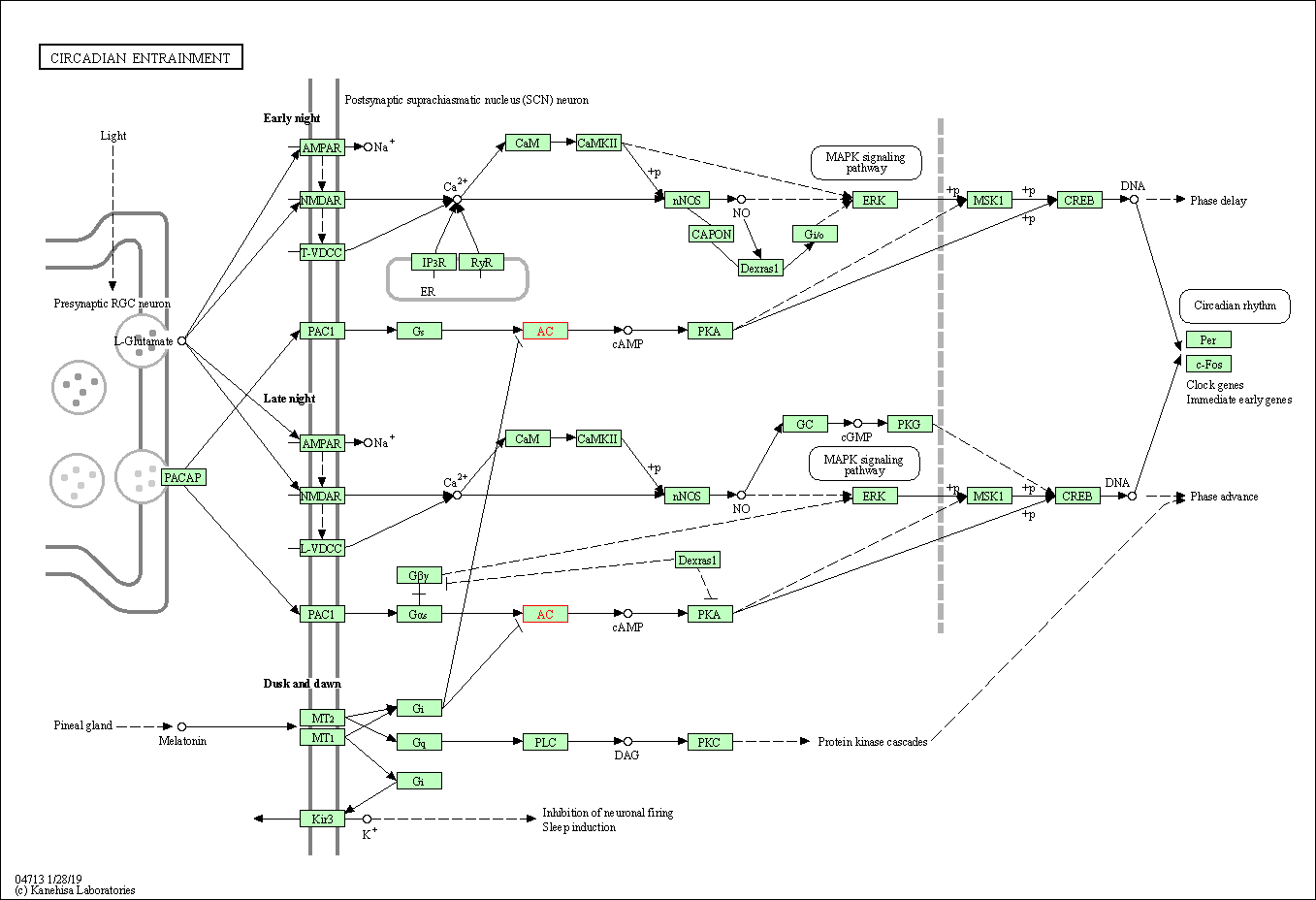
| Circadian entrainment | hsa04713 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
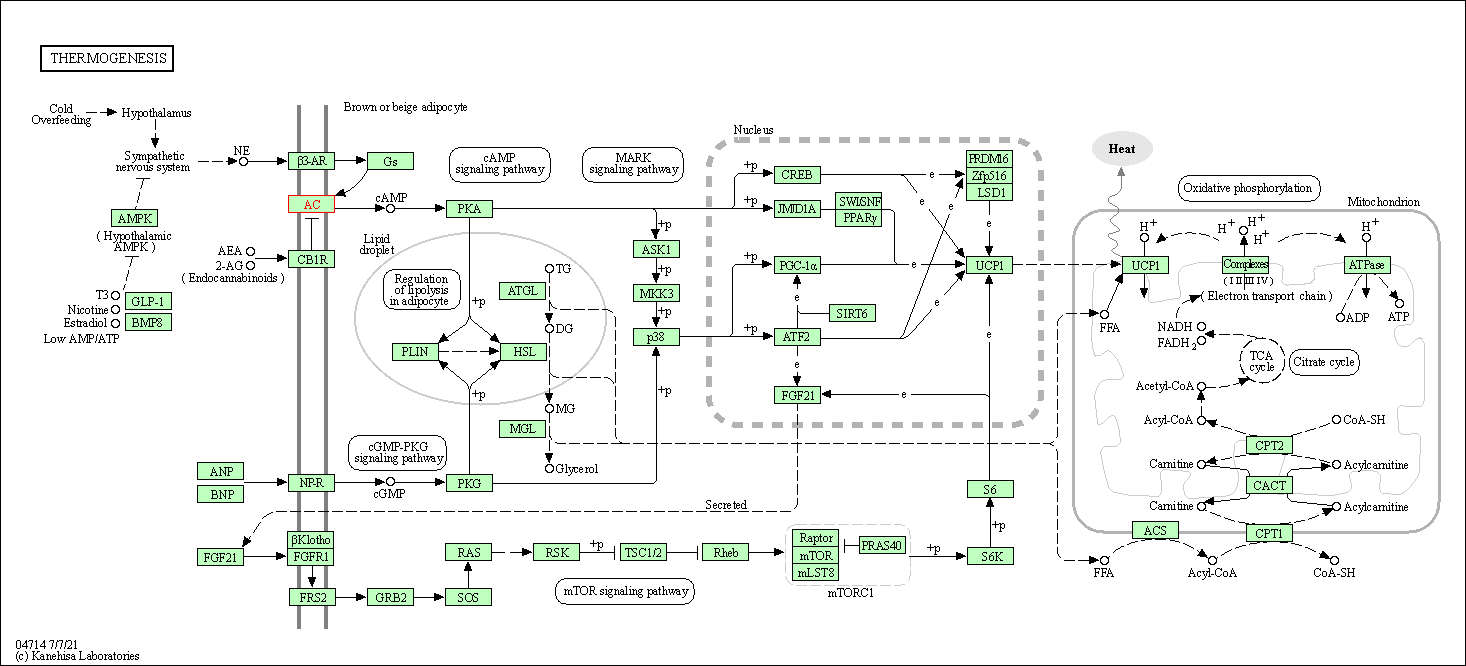
| Thermogenesis | hsa04714 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
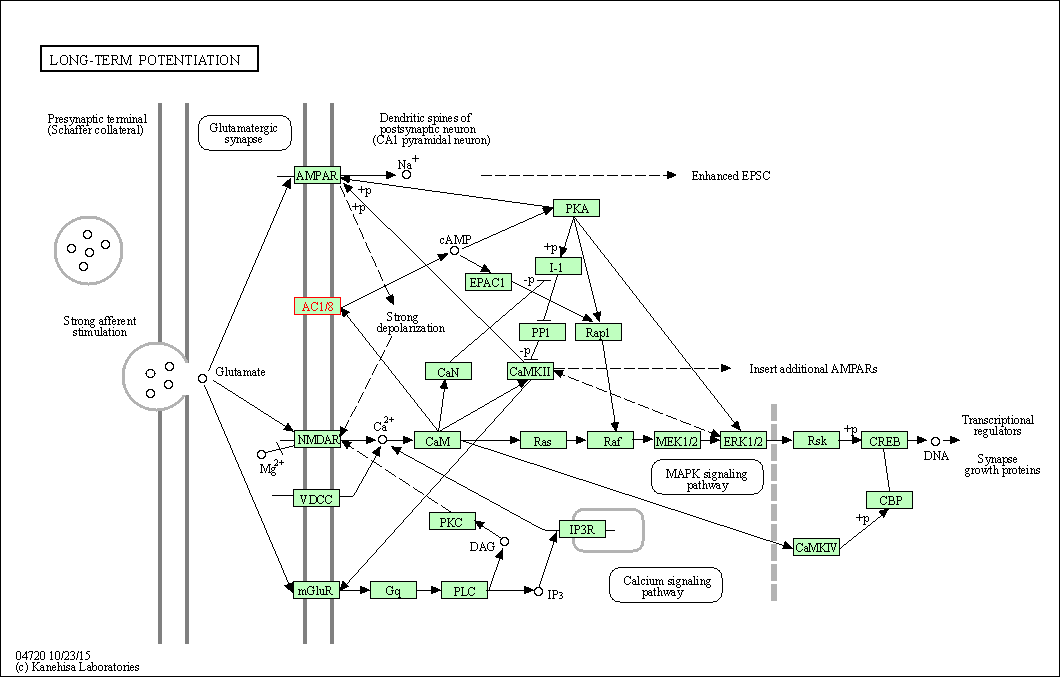
| Long-term potentiation | hsa04720 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Retrograde endocannabinoid signaling | hsa04723 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Glutamatergic synapse | hsa04724 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
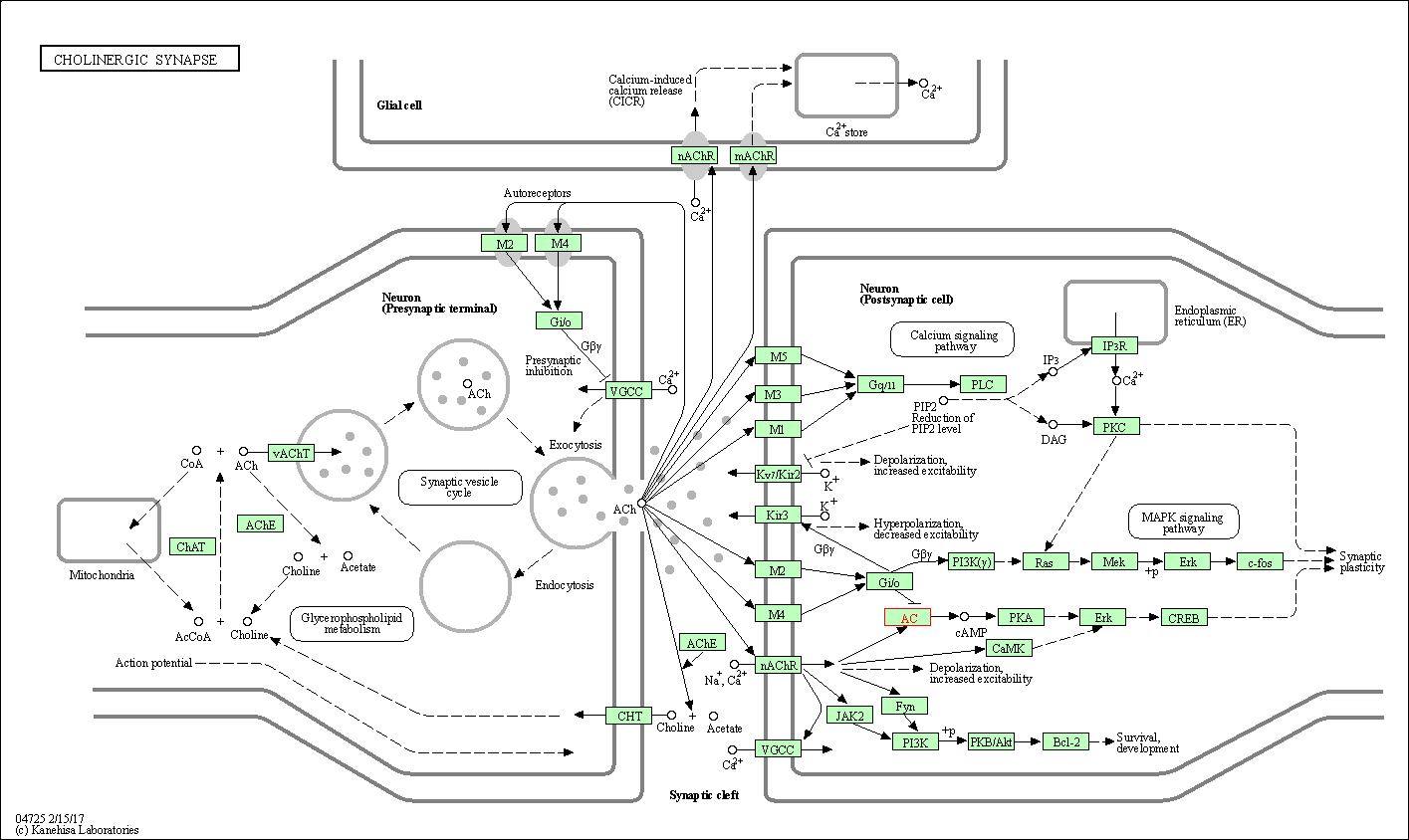
| Cholinergic synapse | hsa04725 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
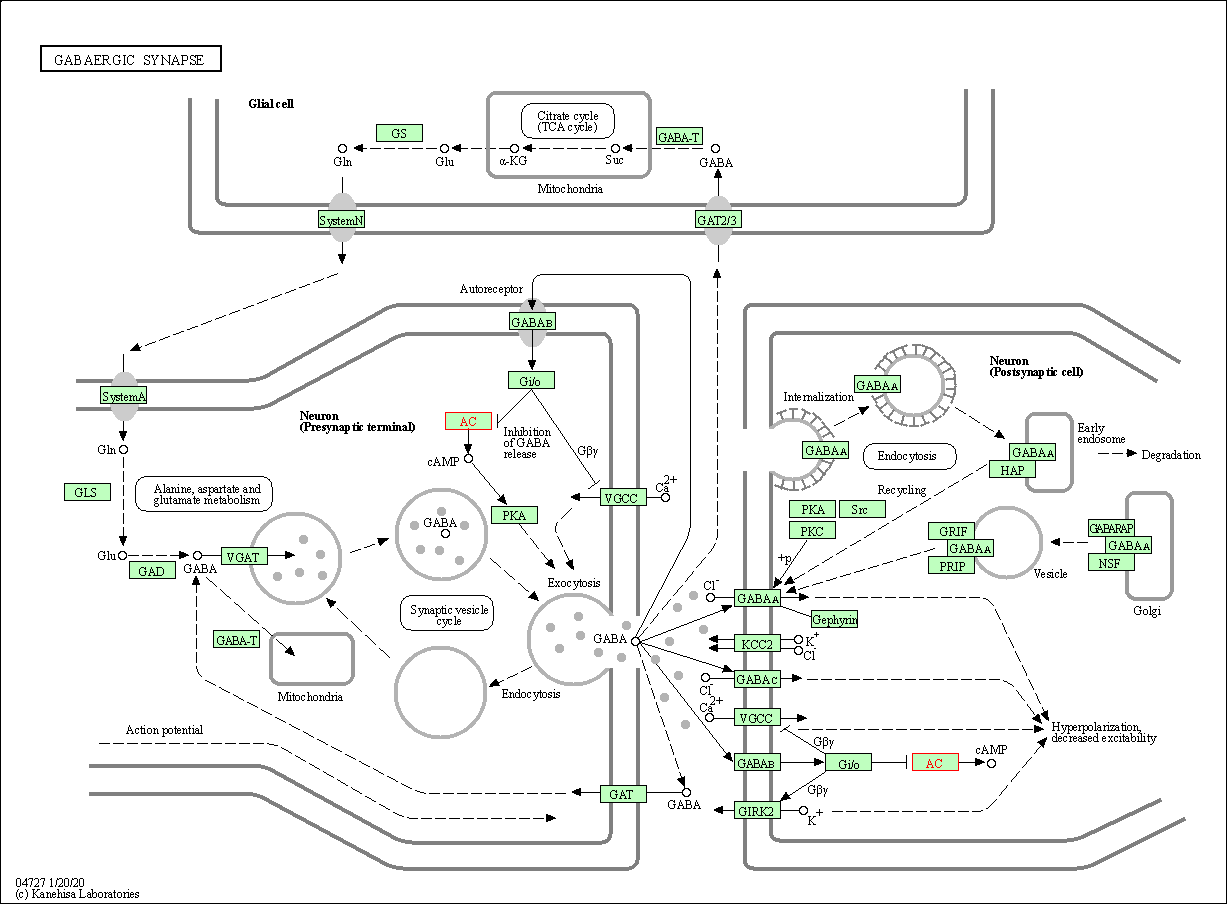
| GABAergic synapse | hsa04727 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
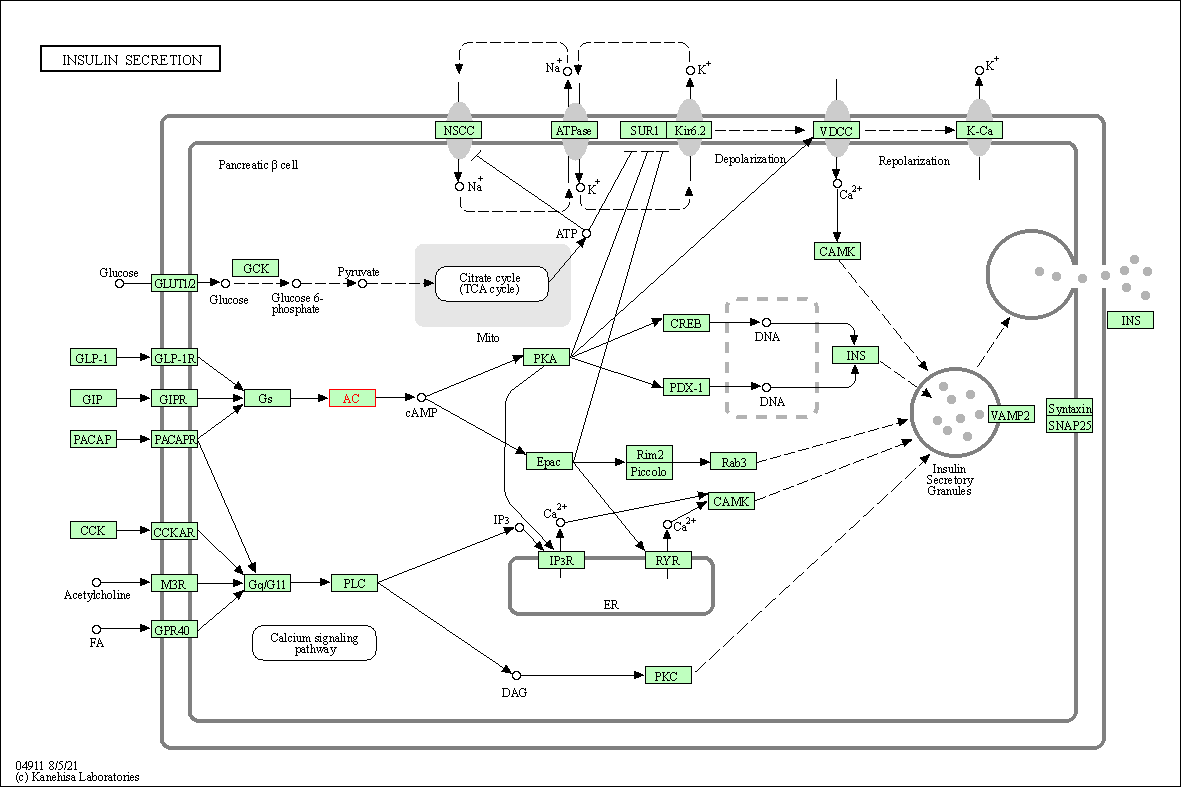
| Insulin secretion | hsa04911 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
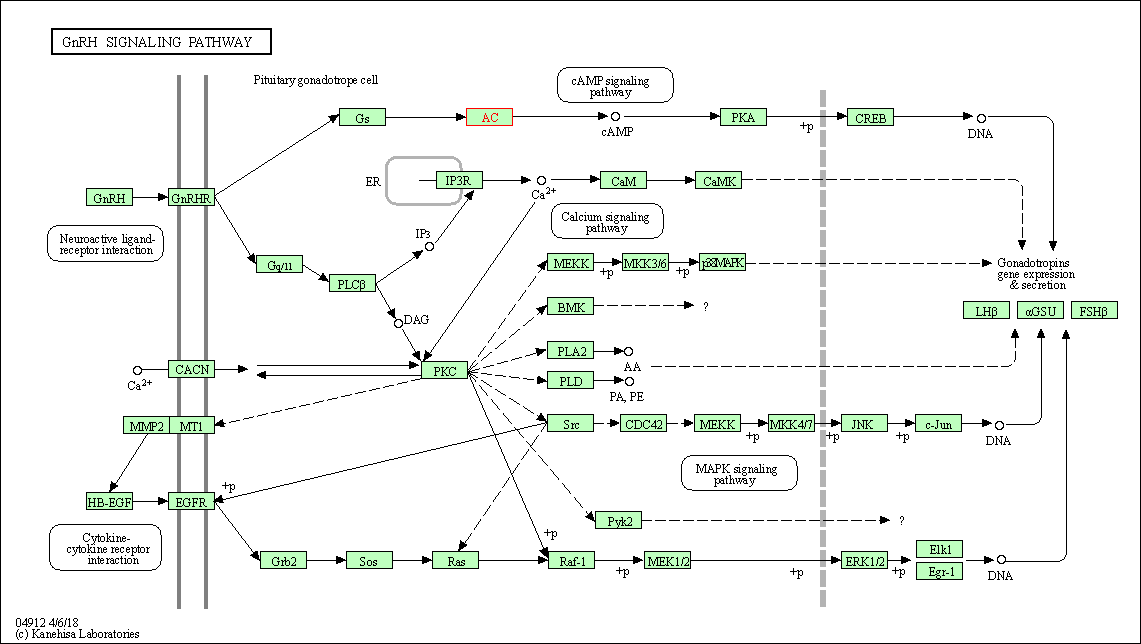
| GnRH signaling pathway | hsa04912 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
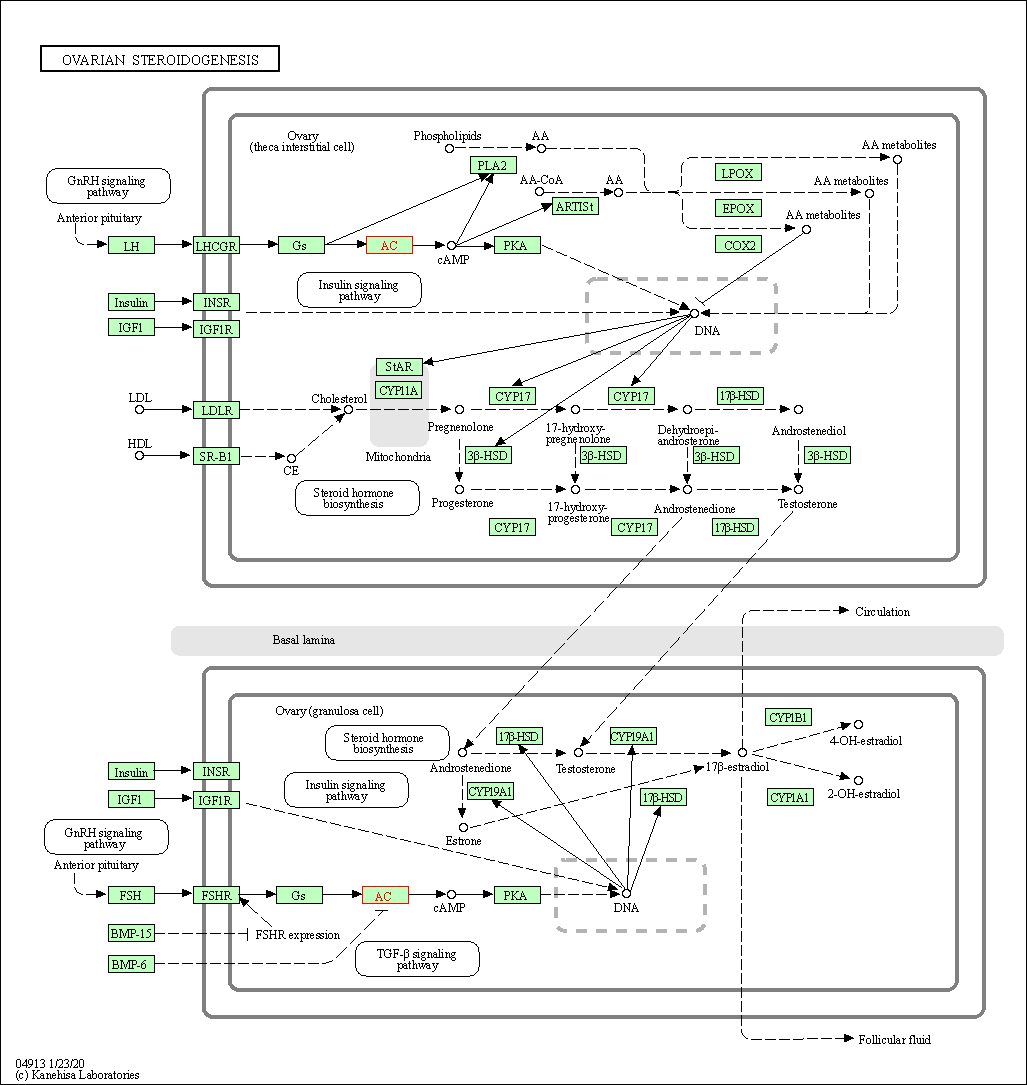
| Ovarian steroidogenesis | hsa04913 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
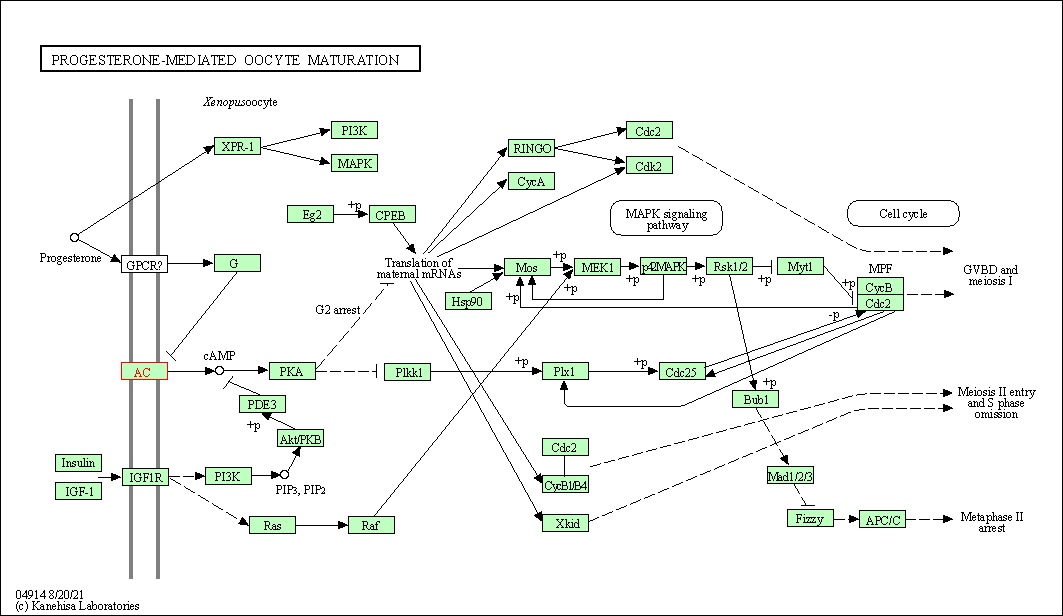
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
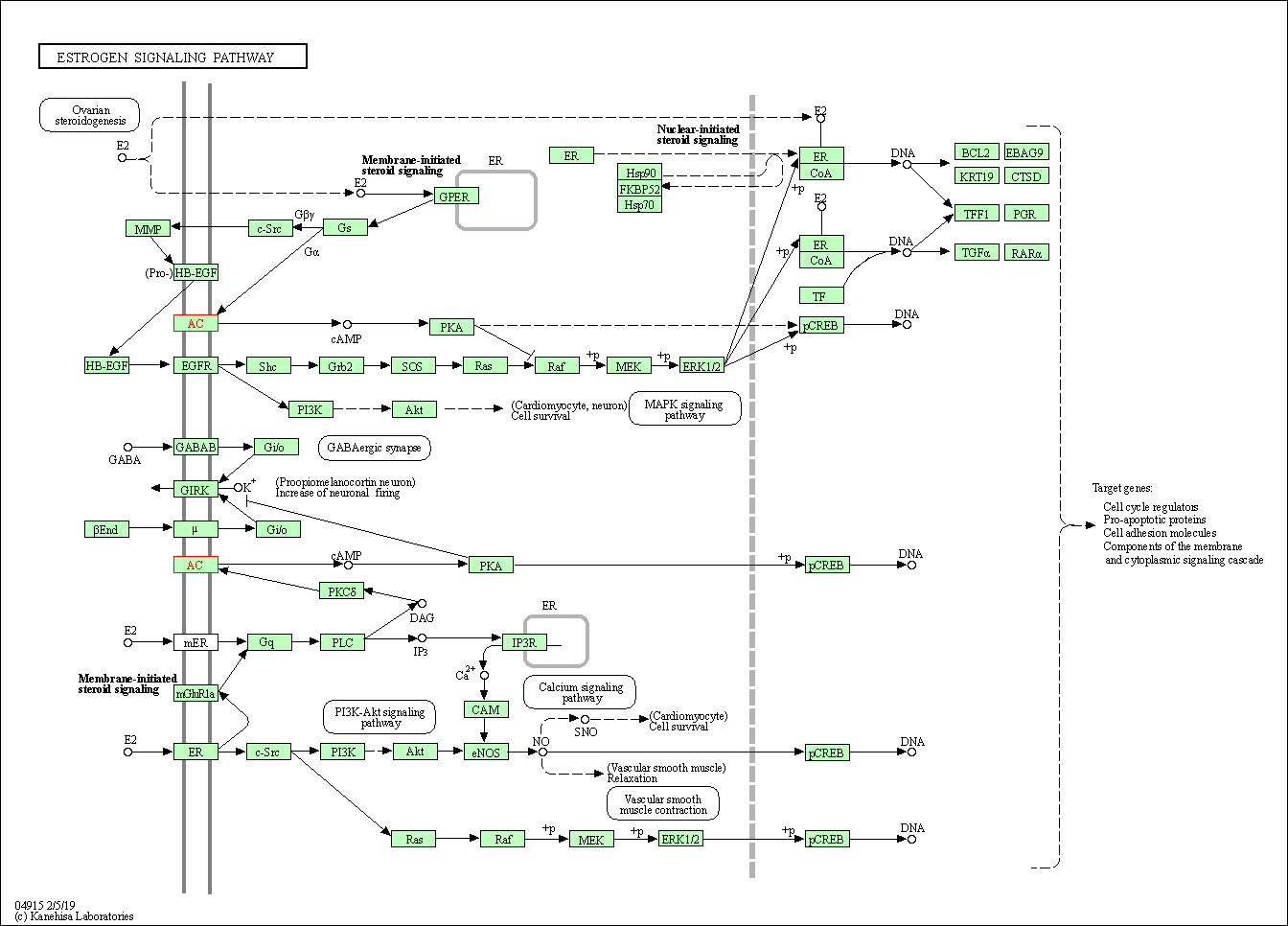
| Estrogen signaling pathway | hsa04915 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Melanogenesis | hsa04916 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone synthesis | hsa04918 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Regulation of lipolysis in adipocytes | hsa04923 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Aldosterone synthesis and secretion | hsa04925 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Cortisol synthesis and secretion | hsa04927 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Parathyroid hormone synthesis, secretion and action | hsa04928 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Salivary secretion | hsa04970 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Gastric acid secretion | hsa04971 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Pancreatic secretion | hsa04972 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Bile secretion | hsa04976 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 3 | Degree centrality | 3.22E-04 | Betweenness centrality | 4.88E-06 |
|---|---|---|---|---|---|
| Closeness centrality | 1.84E-01 | Radiality | 1.31E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 2.00E+01 | Topological coefficient | 4.28E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Ecto- and cytosolic 5'-nucleotidases in normal and AMP deaminase-deficient human skeletal muscle. Biol Chem. 2006 Jan;387(1):53-8. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 2455). | |||||
| REF 3 | Emerging drugs for chemotherapy-induced mucositis. Expert Opin Emerg Drugs. 2008 Sep;13(3):511-22. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

