Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T79694
(Former ID: TTDR00048)
|
|||||
| Target Name |
Voltage-gated calcium channel alpha Cav3.3 (CACNA1I)
|
|||||
| Synonyms |
Voltage-gated calcium channel subunit alpha Cav3.3; Voltage-dependent T-type calcium channel subunit alpha-1I; KIAA1120; Ca(v)3.3
Click to Show/Hide
|
|||||
| Gene Name |
CACNA1I
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Voltage-sensitive calcium channels (VSCC) mediate the entry of calcium ions into excitable cells and are also involved in a variety of calcium-dependent processes, including muscle contraction, hormone or neurotransmitter release, gene expression, cell motility, cell division and cell death. This channel gives rise to T-type calcium currents. T-type calcium channels belong to the "low-voltage activated (LVA)" group and are strongly blocked by nickel and mibefradil. A particularity of this type of channels is an opening at quite negative potentials, and a voltage-dependent inactivation. T-type channels serve pacemaking functions in both central neurons and cardiac nodal cells and support calcium signaling in secretory cells and vascular smooth muscle. They may also be involved in the modulation of firing patterns of neurons which is important for information processing as well as in cell growth processes. Gates in voltage ranges similar to, but higher than alpha 1G or alpha 1H (By similarity).
Click to Show/Hide
|
|||||
| BioChemical Class |
Voltage-gated ion channel
|
|||||
| UniProt ID | ||||||
| Sequence |
MAESASPPSSSAAAPAAEPGVTTEQPGPRSPPSSPPGLEEPLDGADPHVPHPDLAPIAFF
CLRQTTSPRNWCIKMVCNPWFECVSMLVILLNCVTLGMYQPCDDMDCLSDRCKILQVFDD FIFIFFAMEMVLKMVALGIFGKKCYLGDTWNRLDFFIVMAGMVEYSLDLQNINLSAIRTV RVLRPLKAINRVPSMRILVNLLLDTLPMLGNVLLLCFFVFFIFGIIGVQLWAGLLRNRCF LEENFTIQGDVALPPYYQPEEDDEMPFICSLSGDNGIMGCHEIPPLKEQGRECCLSKDDV YDFGAGRQDLNASGLCVNWNRYYNVCRTGSANPHKGAINFDNIGYAWIVIFQVITLEGWV EIMYYVMDAHSFYNFIYFILLIIVGSFFMINLCLVVIATQFSETKQREHRLMLEQRQRYL SSSTVASYAEPGDCYEEIFQYVCHILRKAKRRALGLYQALQSRRQALGPEAPAPAKPGPH AKEPRHYHGKTKGQGDEGRHLGSRHCQTLHGPASPGNDHSGRELCPQHSPLDATPHTLVQ PIPATLASDPASCPCCQHEDGRRPSGLGSTDSGQEGSGSGSSAGGEDEADGDGARSSEDG ASSELGKEEEEEEQADGAVWLCGDVWRETRAKLRGIVDSKYFNRGIMMAILVNTVSMGIE HHEQPEELTNILEICNVVFTSMFALEMILKLAAFGLFDYLRNPYNIFDSIIVIISIWEIV GQADGGLSVLRTFRLLRVLKLVRFMPALRRQLVVLMKTMDNVATFCMLLMLFIFIFSILG MHIFGCKFSLRTDTGDTVPDRKNFDSLLWAIVTVFQILTQEDWNVVLYNGMASTSPWASL YFVALMTFGNYVLFNLLVAILVEGFQAEGDANRSYSDEDQSSSNIEEFDKLQEGLDSSGD PKLCPIPMTPNGHLDPSLPLGGHLGPAGAAGPAPRLSLQPDPMLVALGSRKSSVMSLGRM SYDQRSLSSSRSSYYGPWGRSAAWASRRSSWNSLKHKPPSAEHESLLSAERGGGARVCEV AADEGPPRAAPLHTPHAHHIHHGPHLAHRHRHHRRTLSLDNRDSVDLAELVPAVGAHPRA AWRAAGPAPGHEDCNGRMPSIAKDVFTKMGDRGDRGEDEEEIDYTLCFRVRKMIDVYKPD WCEVREDWSVYLFSPENRFRVLCQTIIAHKLFDYVVLAFIFLNCITIALERPQIEAGSTE RIFLTVSNYIFTAIFVGEMTLKVVSLGLYFGEQAYLRSSWNVLDGFLVFVSIIDIVVSLA SAGGAKILGVLRVLRLLRTLRPLRVISRAPGLKLVVETLISSLKPIGNIVLICCAFFIIF GILGVQLFKGKFYHCLGVDTRNITNRSDCMAANYRWVHHKYNFDNLGQALMSLFVLASKD GWVNIMYNGLDAVAVDQQPVTNHNPWMLLYFISFLLIVSFFVLNMFVGVVVENFHKCRQH QEAEEARRREEKRLRRLEKKRRKAQRLPYYATYCHTRLLIHSMCTSHYLDIFITFIICLN VVTMSLEHYNQPTSLETALKYCNYMFTTVFVLEAVLKLVAFGLRRFFKDRWNQLDLAIVL LSVMGITLEEIEINAALPINPTIIRIMRVLRIARVLKLLKMATGMRALLDTVVQALPQVG NLGLLFMLLFFIYAALGVELFGKLVCNDENPCEGMSRHATFENFGMAFLTLFQVSTGDNW NGIMKDTLRDCTHDERSCLSSLQFVSPLYFVSFVLTAQFVLINVVVAVLMKHLDDSNKEA QEDAEMDAELELEMAHGLGPGPRLPTGSPGAPGRGPGGAGGGGDTEGGLCRRCYSPAQEN LWLDSVSLIIKDSLEGELTIIDNLSGSIFHHYSSPAGCKKCHHDKQEVQLAETEAFSLNS DRSSSILLGDDLSLEDPTACPPGRKDSKGELDPPEPMRVGDLGECFFPLSSTAVSPDPEN FLCEMEEIPFNPVRSWLKHDSSQAPPSPFSPDASSPLLPMPAEFFHPAVSASQKGPEKGT GTGTLPKIALQGSWASLRSPRVNCTLLRQATGSDTSLDASPSSSAGSLQTTLEDSLTLSD SPRRALGPPAPAPGPRAGLSPAARRRLSLRGRGLFSLRGLRAHQRSHSSGGSTSPGCTHH DSMDPSDEEGRGGAGGGGAGSEHSETLSSLSLTSLFCPPPPPPAPGLTPARKFSSTSSLA APGRPHAAALAHGLARSPSWAADRSKDPPGRAPLPMGLGPLAPPPQPLPGELEPGDAASK RKR Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T38L2B | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Pimozide | Ligand Info | |||||
| Structure Description | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with pimozide(PMZ) | PDB:7WLL | ||||
| Method | Electron microscopy | Resolution | 3.60 Å | Mutation | No | [3] |
| PDB Sequence |
TSPRNWCIKM
75 VCNPWFECVS85 MLVILLNCVT95 LGMYQPCDDM105 DCLSDRCKIL115 QVFDDFIFIF 125 FAMEMVLKMV135 ALGIFGKKCY145 LGDTWNRLDF155 FIVMAGMVEY165 SLDLQNINLS 175 AIRTVRVLRP185 LKAINRVPSM195 RILVNLLLDT205 LPMLGNVLLL215 CFFVFFIFGI 225 IGVQLWAGLL235 RNRCFLEENF245 TIQGDVALPP255 YYQPEEDDEM265 PFICSLSGDN 275 GIMGCHEIPP285 LKCVNWNRYY323 NVCRTGSANP333 HKGAINFDNI343 GYAWIVIFQV 353 ITLEGWVEIM363 YYVMDAHSFY373 NFIYFILLII383 VGSFFMINLC393 LVVIATQFSE 403 TKQREHRLML413 RETRAKLRGI636 VDSKYFNRGI646 MMAILVNTVS656 MGIEHHEQPE 666 ELTNILEICN676 VVFTSMFALE686 MILKLAAFGL696 FDYLRNPYNI706 FDSIIVIISI 716 WEIVGQADGG726 LSVLRTFRLL736 RVLKLVRFMP746 ALRRQLVVLM756 KTMDNVATFC 766 MLLMLFIFIF776 SILGMHIFGC786 KFSLRTDTGD796 TVPDRKNFDS806 LLWAIVTVFQ 816 ILTQEDWNVV826 LYNGMASTSP836 WASLYFVALM846 TFGNYVLFNL856 LVAILVEGFQ 866 QTIIAHKLFD1173 YVVLAFIFLN1183 CITIALERPQ1193 IEAGSTERIF1203 LTVSNYIFTA 1213 IFVGEMTLKV1223 VSLGLYFGEQ1233 AYLRSSWNVL1243 DGFLVFVSII1253 DIVVSLASAG 1263 GAKILGVLRV1273 LRLLRTLRPL1283 RVISRAPGLK1293 LVVETLISSL1303 KPIGNIVLIC 1313 CAFFIIFGIL1323 GVQLFKGKFY1333 HCLGVDTRNI1343 TNRSDCMAAN1353 YRWVHHKYNF 1363 DNLGQALMSL1373 FVLASKDGWV1383 NIMYNGLDAV1393 AVDQQPVTNH1403 NPWMLLYFIS 1413 FLLIVSFFVL1423 NMFVGVVVEN1433 FHKCRQHQEA1443 EEARRREEKR1453 LRRLEKKRRK 1463 AQRLPYYATY1473 CHTRLLIHSM1483 CTSHYLDIFI1493 TFIICLNVVT1503 MSLEHYNQPT 1513 SLETALKYCN1523 YMFTTVFVLE1533 AVLKLVAFGL1543 RRFFKDRWNQ1553 LDLAIVLLSV 1563 MGITLEEIEI1573 NAALPINPTI1583 IRIMRVLRIA1593 RVLKLLKMAT1603 GMRALLDTVV 1613 QALPQVGNLG1623 LLFMLLFFIY1633 AALGVELFGK1643 LVCNDENPCE1653 GMSRHATFEN 1663 FGMAFLTLFQ1673 VSTGDNWNGI1683 MKDTLRDCTH1693 DERSCLSSLQ1703 FVSPLYFVSF 1713 VLTAQFVLIN1723 VVVAVLMKHL1733 DDSNK
|
|||||
|
|
PHE387
3.961
ASN391
3.320
LEU394
3.724
PHE815
4.315
LEU818
2.962
THR819
3.809
GLN820
3.959
ASN850
2.026
PHE854
3.337
VAL858
3.470
LYS1379
3.588
|
|||||
| Ligand Name: Mibefradil | Ligand Info | |||||
| Structure Description | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with mibefradil (MIB) | PDB:7WLJ | ||||
| Method | Electron microscopy | Resolution | 3.90 Å | Mutation | No | [3] |
| PDB Sequence |
TSPRNWCIKM
75 VCNPWFECVS85 MLVILLNCVT95 LGMYQPCDDM105 DCLSDRCKIL115 QVFDDFIFIF 125 FAMEMVLKMV135 ALGIFGKKCY145 LGDTWNRLDF155 FIVMAGMVEY165 SLDLQNINLS 175 AIRTVRVLRP185 LKAINRVPSM195 RILVNLLLDT205 LPMLGNVLLL215 CFFVFFIFGI 225 IGVQLWAGLL235 RNRCFLEENF245 TIQGDVALPP255 YYQPEEDDEM265 PFICSLSGDN 275 GIMGCHEIPP285 LKCVNWNRYY323 NVCRTGSANP333 HKGAINFDNI343 GYAWIVIFQV 353 ITLEGWVEIM363 YYVMDAHSFY373 NFIYFILLII383 VGSFFMINLC393 LVVIATQFSE 403 TKQREHRLML413 RETRAKLRGI636 VDSKYFNRGI646 MMAILVNTVS656 MGIEHHEQPE 666 ELTNILEICN676 VVFTSMFALE686 MILKLAAFGL696 FDYLRNPYNI706 FDSIIVIISI 716 WEIVGQADGG726 LSVLRTFRLL736 RVLKLVRFMP746 ALRRQLVVLM756 KTMDNVATFC 766 MLLMLFIFIF776 SILGMHIFGC786 KFSLRTDTGD796 TVPDRKNFDS806 LLWAIVTVFQ 816 ILTQEDWNVV826 LYNGMASTSP836 WASLYFVALM846 TFGNYVLFNL856 LVAILVEGFQ 866 QTIIAHKLFD1173 YVVLAFIFLN1183 CITIALERPQ1193 IEAGSTERIF1203 LTVSNYIFTA 1213 IFVGEMTLKV1223 VSLGLYFGEQ1233 AYLRSSWNVL1243 DGFLVFVSII1253 DIVVSLASAG 1263 GAKILGVLRV1273 LRLLRTLRPL1283 RVISRAPGLK1293 LVVETLISSL1303 KPIGNIVLIC 1313 CAFFIIFGIL1323 GVQLFKGKFY1333 HCLGVDTRNI1343 TNRSDCMAAN1353 YRWVHHKYNF 1363 DNLGQALMSL1373 FVLASKDGWV1383 NIMYNGLDAV1393 AVDQQPVTNH1403 NPWMLLYFIS 1413 FLLIVSFFVL1423 NMFVGVVVEN1433 FHKCRQHQEA1443 EEARRREEKR1453 LRRLEKKRRK 1463 AQRLPYYATY1473 CHTRLLIHSM1483 CTSHYLDIFI1493 TFIICLNVVT1503 MSLEHYNQPT 1513 SLETALKYCN1523 YMFTTVFVLE1533 AVLKLVAFGL1543 RRFFKDRWNQ1553 LDLAIVLLSV 1563 MGITLEEIEI1573 NAALPINPTI1583 IRIMRVLRIA1593 RVLKLLKMAT1603 GMRALLDTVV 1613 QALPQVGNLG1623 LLFMLLFFIY1633 AALGVELFGK1643 LVCNDENPCE1653 GMSRHATFEN 1663 FGMAFLTLFQ1673 VSTGDNWNGI1683 MKDTLRDCTH1693 DERSCLSSLQ1703 FVSPLYFVSF 1713 VLTAQFVLIN1723 VVVAVLMKHL1733 DDSNK
|
|||||
|
|
PHE387
3.769
ILE390
4.075
ASN391
3.308
LEU394
3.558
PHE815
3.451
LEU818
3.427
THR819
3.785
GLN820
4.063
ASN850
2.881
PHE854
3.532
ASN855
4.597
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|


| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
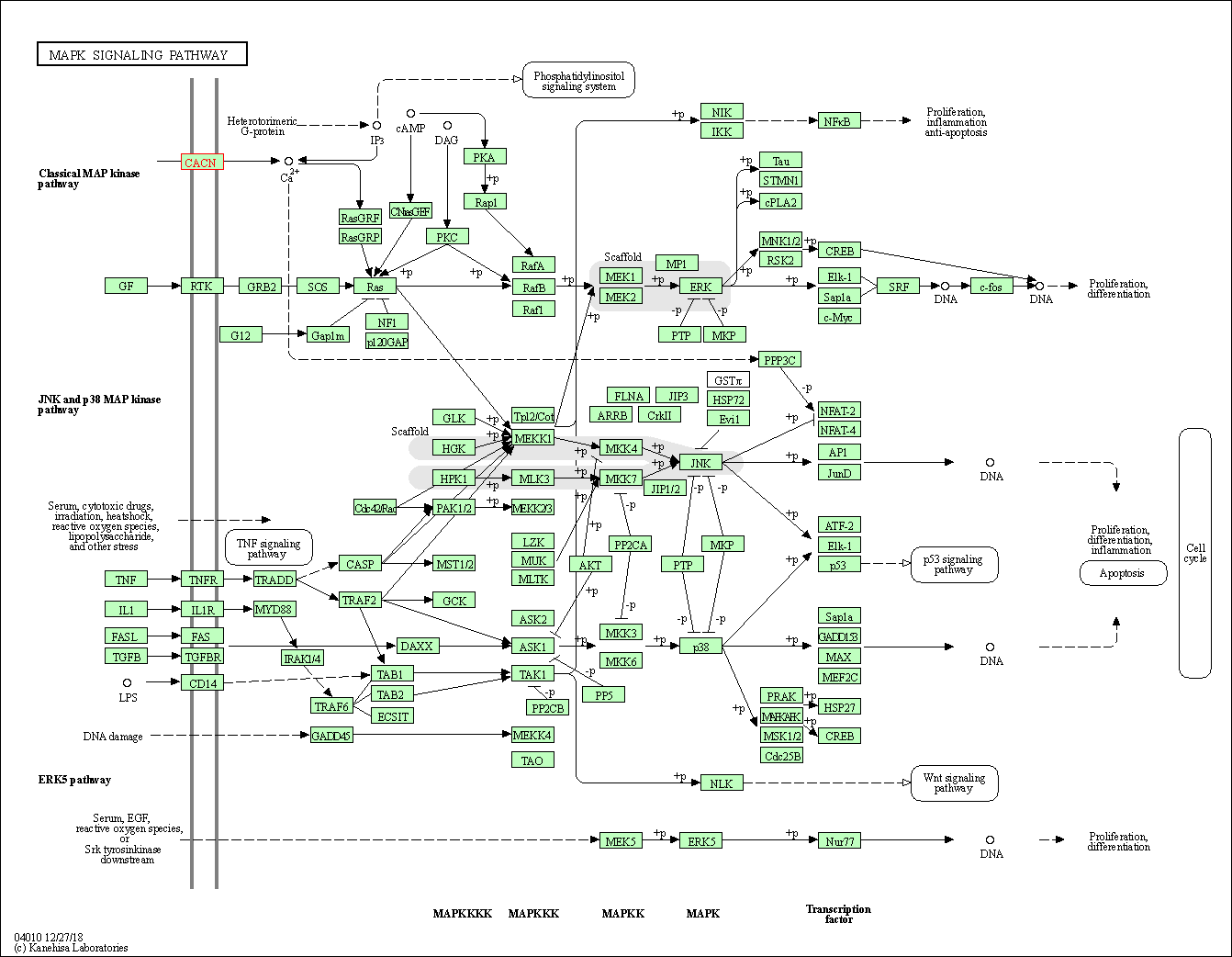
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
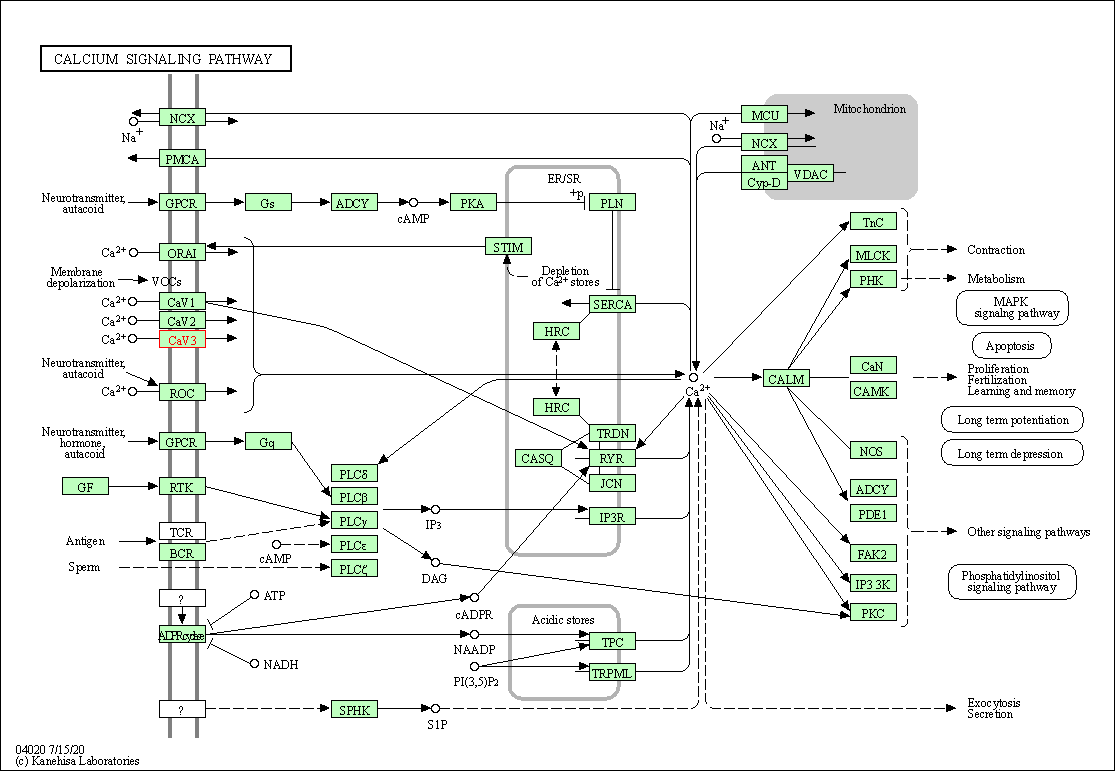
| Calcium signaling pathway | hsa04020 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Circadian entrainment | hsa04713 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
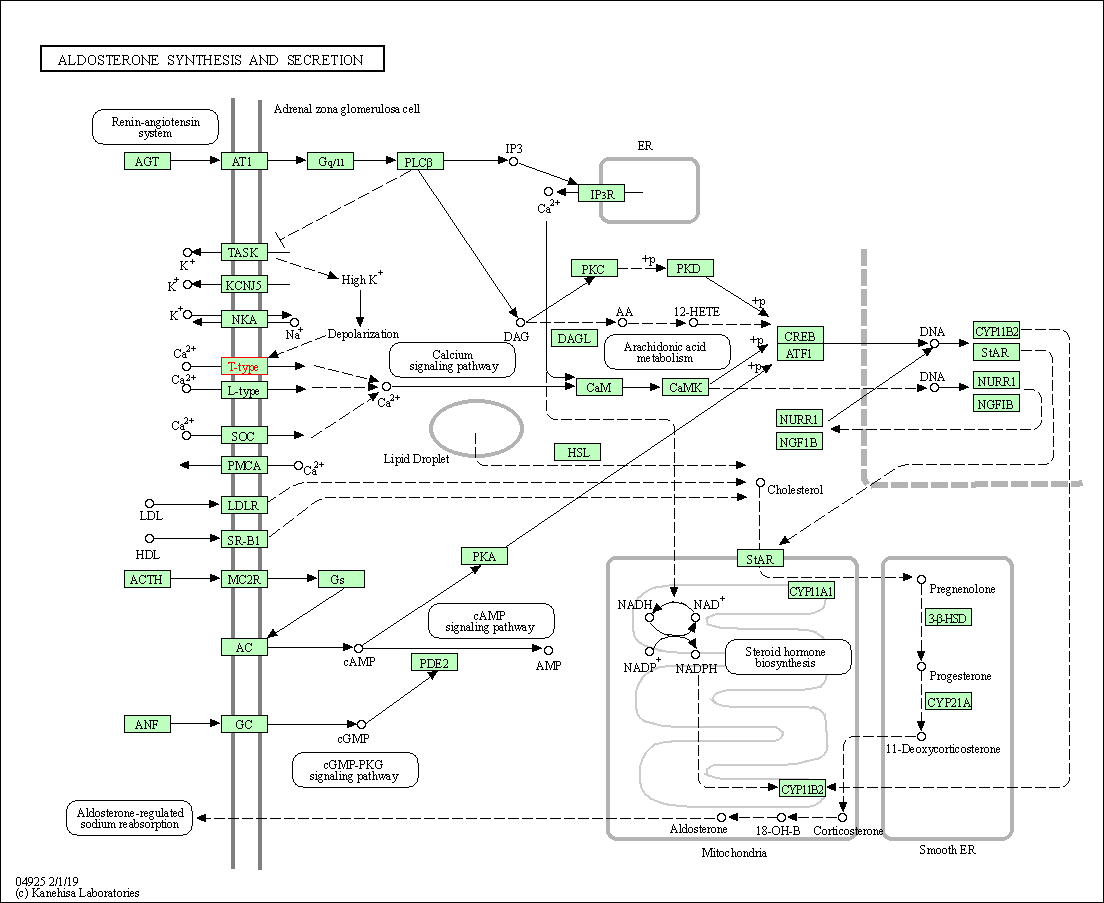
| Aldosterone synthesis and secretion | hsa04925 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Cortisol synthesis and secretion | hsa04927 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
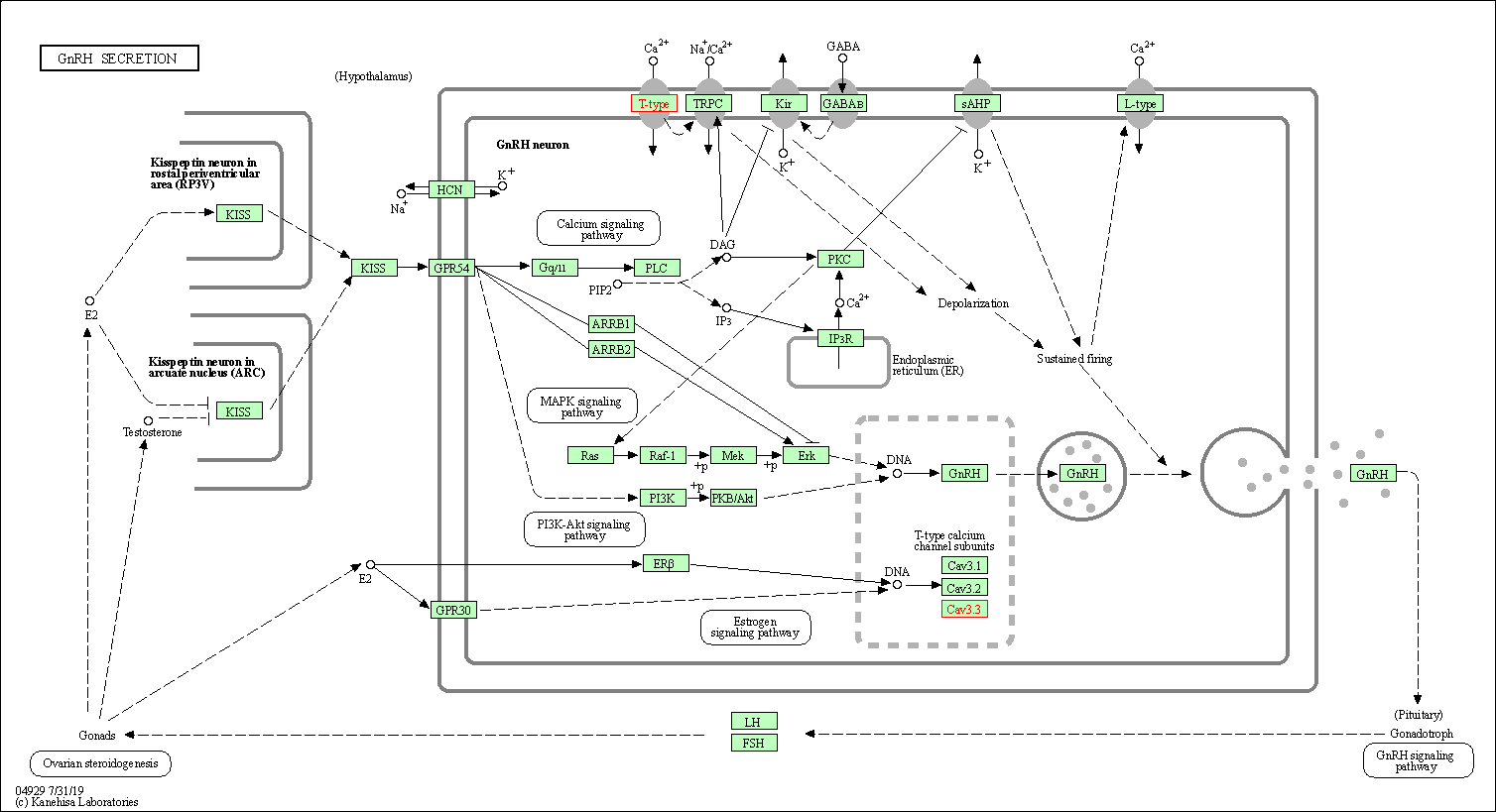
| GnRH secretion | hsa04929 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | MAPK signaling pathway | |||||
| 2 | Calcium signaling pathway | |||||
| 3 | Circadian entrainment | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Discovery of (2S)-1-(4-amino-2,3,5- trimethylphenoxy)-3-[4-[4-(4- fluorobenzyl)phenyl]-1-piperazinyl]-2-propanol dimethanesulfonate (SUN N8075): a ... J Med Chem. 2000 Sep 7;43(18):3372-6. | |||||
| REF 2 | The Discovery and Characterization of ML218: A Novel, Centrally Active T-Type Calcium Channel Inhibitor with Robust Effects in STN Neurons and in a Rodent Model of Parkinson's Disease. ACS Chem Neurosci. 2011 Dec 21;2(12):730-742. | |||||
| REF 3 | Structure, gating, and pharmacology of human Ca(V)3.3 channel. Nat Commun. 2022 Apr 19;13(1):2084. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

