Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T83797
(Former ID: TTDC00194)
|
|||||
| Target Name |
Hepatocyte growth factor (HGF)
|
|||||
| Synonyms |
Scatter factor; SF; Hepatopoietin-A; Hepatopoeitin A; HPTA
Click to Show/Hide
|
|||||
| Gene Name |
HGF
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 5 Target-related Diseases | + | ||||
| 1 | Angina pectoris [ICD-11: BA40] | |||||
| 2 | Chronic arterial occlusive disease [ICD-11: BD4Z] | |||||
| 3 | Diabetic foot ulcer [ICD-11: BD54] | |||||
| 4 | Graft-versus-host disease [ICD-11: 4B24] | |||||
| 5 | Neuropathy [ICD-11: 8C0Z] | |||||
| Function |
Activating ligand for the receptor tyrosine kinase MET by binding to it and promoting its dimerization. Potent mitogen for mature parenchymal hepatocyte cells, seems to be a hepatotrophic factor, and acts as a growth factor for a broad spectrum of tissues and cell types.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| Sequence |
MWVTKLLPALLLQHVLLHLLLLPIAIPYAEGQRKRRNTIHEFKKSAKTTLIKIDPALKIK
TKKVNTADQCANRCTRNKGLPFTCKAFVFDKARKQCLWFPFNSMSSGVKKEFGHEFDLYE NKDYIRNCIIGKGRSYKGTVSITKSGIKCQPWSSMIPHEHSFLPSSYRGKDLQENYCRNP RGEEGGPWCFTSNPEVRYEVCDIPQCSEVECMTCNGESYRGLMDHTESGKICQRWDHQTP HRHKFLPERYPDKGFDDNYCRNPDGQPRPWCYTLDPHTRWEYCAIKTCADNTMNDTDVPL ETTECIQGQGEGYRGTVNTIWNGIPCQRWDSQYPHEHDMTPENFKCKDLRENYCRNPDGS ESPWCFTTDPNIRVGYCSQIPNCDMSHGQDCYRGNGKNYMGNLSQTRSGLTCSMWDKNME DLHRHIFWEPDASKLNENYCRNPDDDAHGPWCYTGNPLIPWDYCPISRCEGDTTPTIVNL DHPVISCAKTKQLRVVNGIPTRTNIGWMVSLRYRNKHICGGSLIKESWVLTARQCFPSRD LKDYEAWLGIHDVHGRGDEKCKQVLNVSQLVYGPEGSDLVLMKLARPAVLDDFVSTIDLP NYGCTIPEKTSCSVYGWGYTGLINYDGLLRVAHLYIMGNEKCSQHHRGKVTLNESEICAG AEKIGSGPCEGDYGGPLVCEQHKMRMVLGVIVPGRGCAIPNRPGIFVRVAYYAKWIHKII LTYKVPQS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A02005 | |||||
| HIT2.0 ID | T45X9Z | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 8 Clinical Trial Drugs | + | ||||
| 1 | ANG-3777 | Drug Info | Phase 3 | Delayed graft function | [2] | |
| 2 | Donaperminogene seltoplasmid | Drug Info | Phase 3 | Diabetic foot ulcer | [3] | |
| 3 | VM-202 | Drug Info | Phase 3 | Angina pectoris | [4] | |
| 4 | Collategene | Drug Info | Phase 2 | Peripheral Ischemic Ulcers | [5] | |
| 5 | Ficlatuzumab | Drug Info | Phase 2 | Non-small-cell lung cancer | [6] | |
| 6 | MP0250 | Drug Info | Phase 1/2 | Non-small-cell lung cancer | [7] | |
| 7 | ABI-011 | Drug Info | Phase 1 | Solid tumour/cancer | [8] | |
| 8 | TAK-701 | Drug Info | Phase 1 | Advanced malignancy | [9] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | Rilotumumab | Drug Info | Discontinued in Phase 2 | Grade IV malignant glioma | [10], [11] | |
| Mode of Action | [+] 4 Modes of Action | + | ||||
| Mimetic | [+] 1 Mimetic drugs | + | ||||
| 1 | ANG-3777 | Drug Info | [12] | |||
| Replacement | [+] 1 Replacement drugs | + | ||||
| 1 | Donaperminogene seltoplasmid | Drug Info | [13] | |||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | VM-202 | Drug Info | [1] | |||
| 2 | TAK-701 | Drug Info | [15] | |||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Ficlatuzumab | Drug Info | [7] | |||
| 2 | MP0250 | Drug Info | [7] | |||
| 3 | ABI-011 | Drug Info | [7] | |||
| 4 | N,O6-Disulfo-Glucosamine | Drug Info | [18] | |||
| 5 | O2-Sulfo-Glucuronic Acid | Drug Info | [18] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: N,O6-Disulfo-Glucosamine | Ligand Info | |||||
| Structure Description | Crystal structure of 1K1 mutant of Hepatocyte Growth Factor/Scatter Factor fragment NK1 in complex with heparin | PDB:3MKP | ||||
| Method | X-ray diffraction | Resolution | 2.81 Å | Mutation | Yes | [19] |
| PDB Sequence |
RNTIHEFKKS
45 AKTTLIKIDP55 ALKIKTKKVN65 TADQCANRCT75 RNKGLPFTCK85 AFVFDKARKQ 95 CLWFPFNSMS105 SGVKKEFGHE115 FDLYENKDYI125 RNCIIGEGES135 YKGTVSITKS 145 GIKCQPWSSM155 IPHEHSFLPS165 SYRGKDLQEN175 YCRNPRGEEG185 GPWCFTSNPE 195 VRYEVCDIPQ205 CSE
|
|||||
|
|
||||||
| Ligand Name: 3[N-Morpholino]propane sulfonic acid | Ligand Info | |||||
| Structure Description | The structure of the NK1 fragment of HGF/SF complexed with MOPS | PDB:5CSQ | ||||
| Method | X-ray diffraction | Resolution | 1.95 Å | Mutation | Yes | [20] |
| PDB Sequence |
TIHEFKKSAK
47 TTLIKIDPAL57 KIKTKKVNTA67 DQCANRCTRN77 KGLPFTCKAF87 VFDKARKQCL 97 WFPFNSMSSG107 VKKEFGHEFD117 LYENKDYIRN127 CIIGKGRSYK137 GTVSITKSGI 147 KCQPWSSMIP157 HEHSFLPSSY167 RGKDLQENYC177 RNPRGEEGGP187 WCFTSNPEVR 197 YEVCDIPQCS207 EVE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
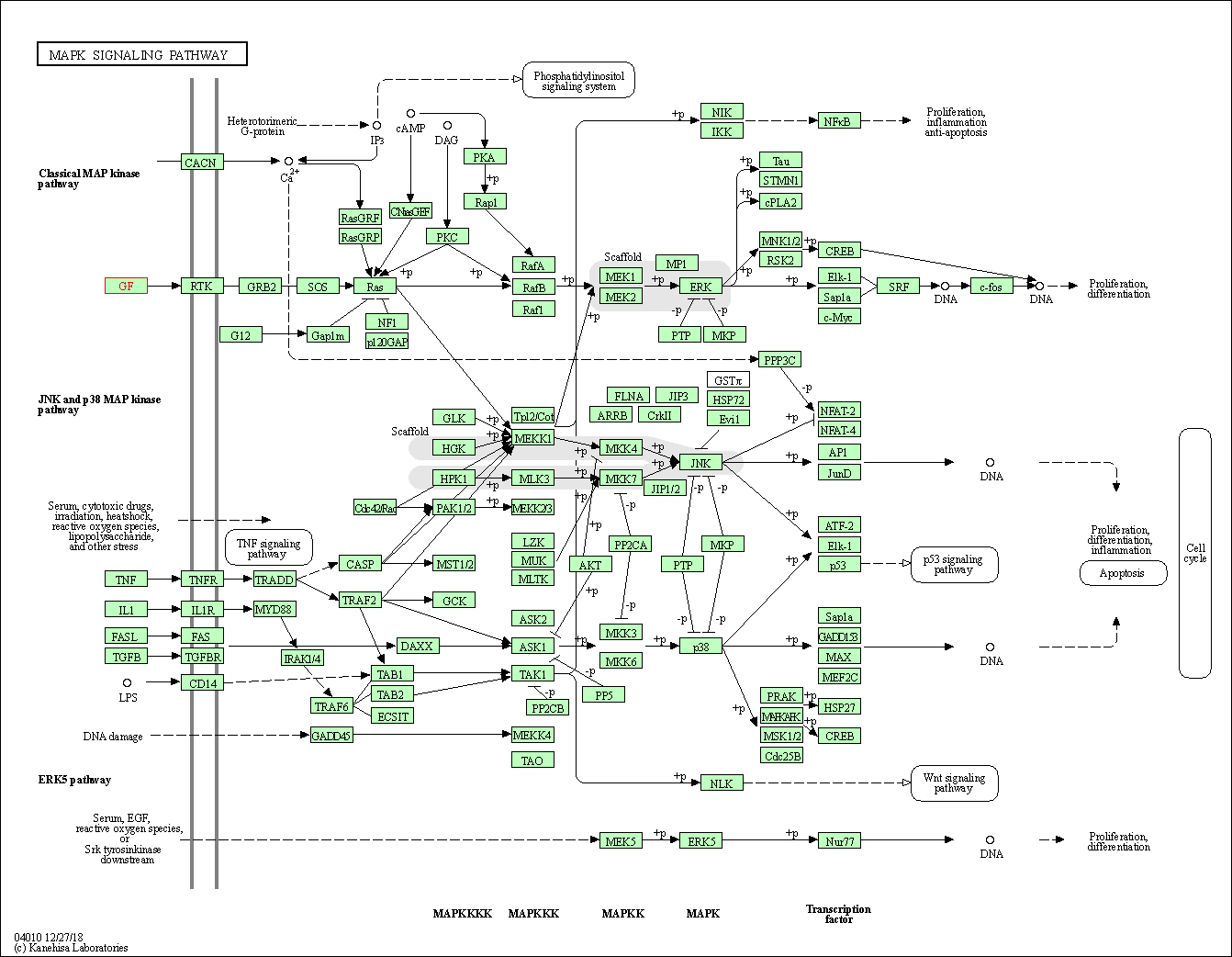
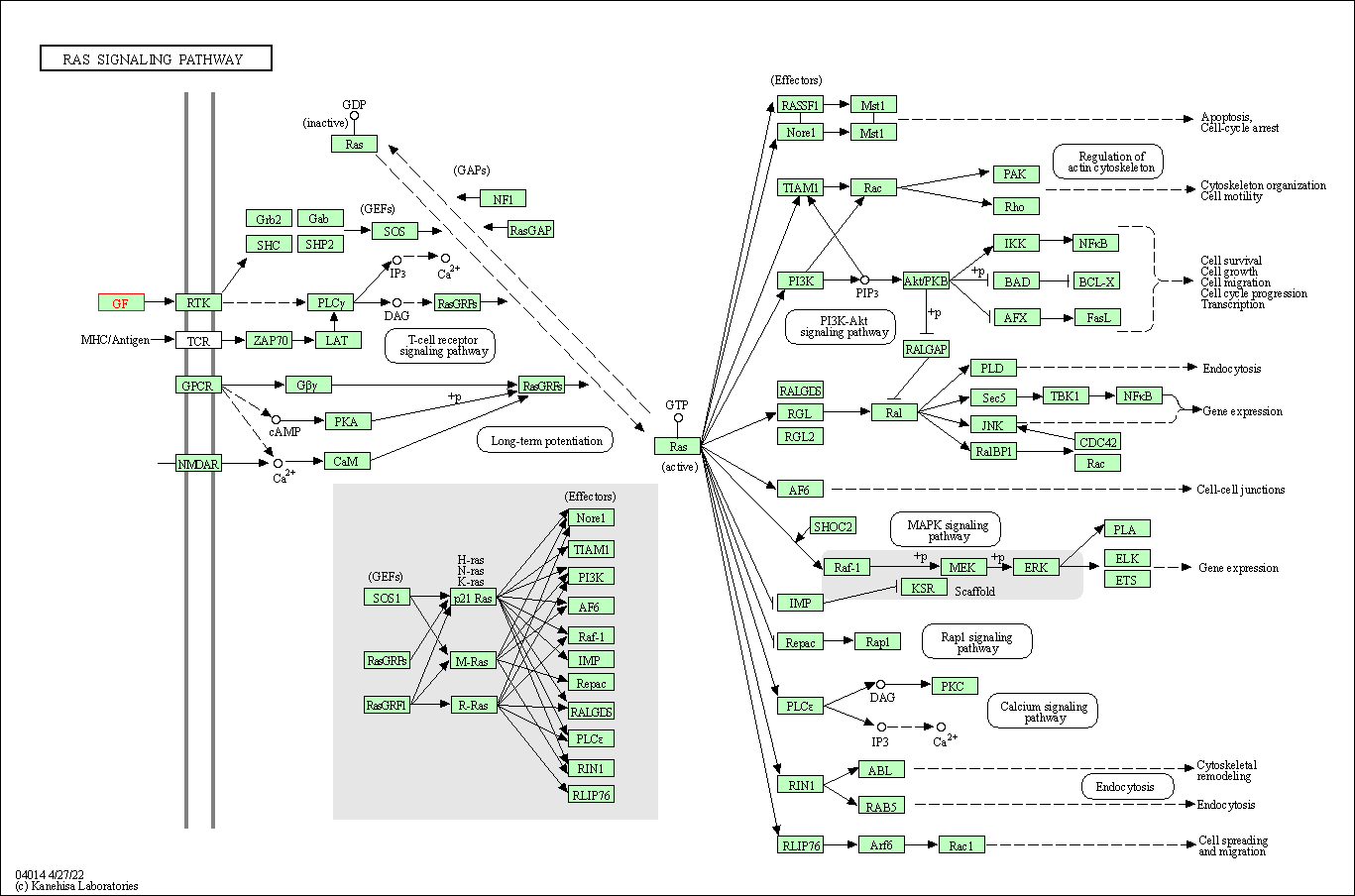
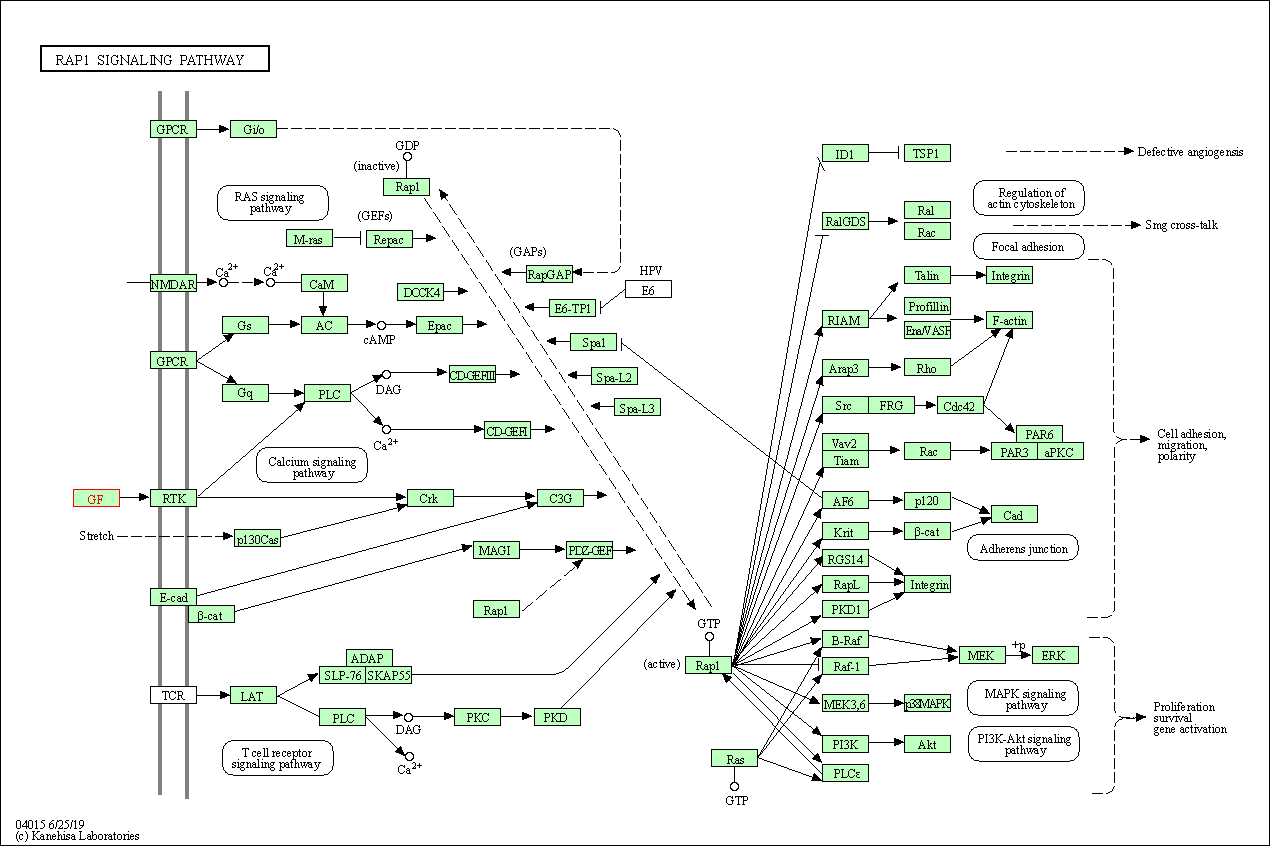
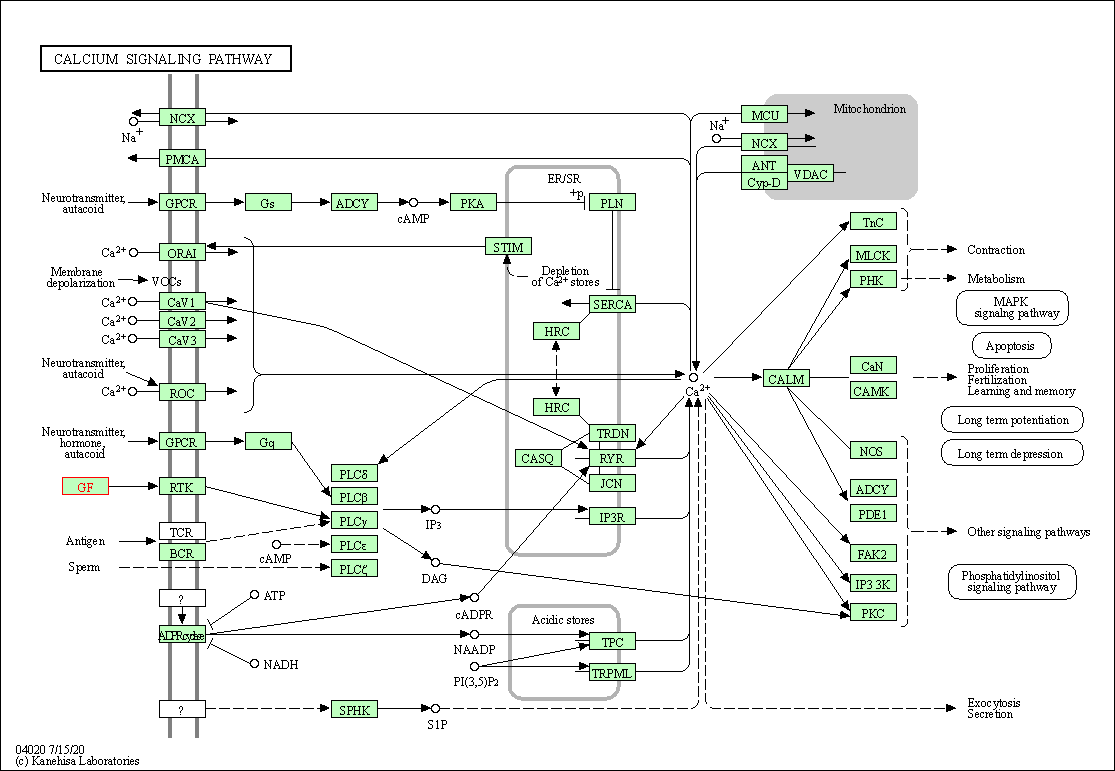
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Calcium signaling pathway | hsa04020 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
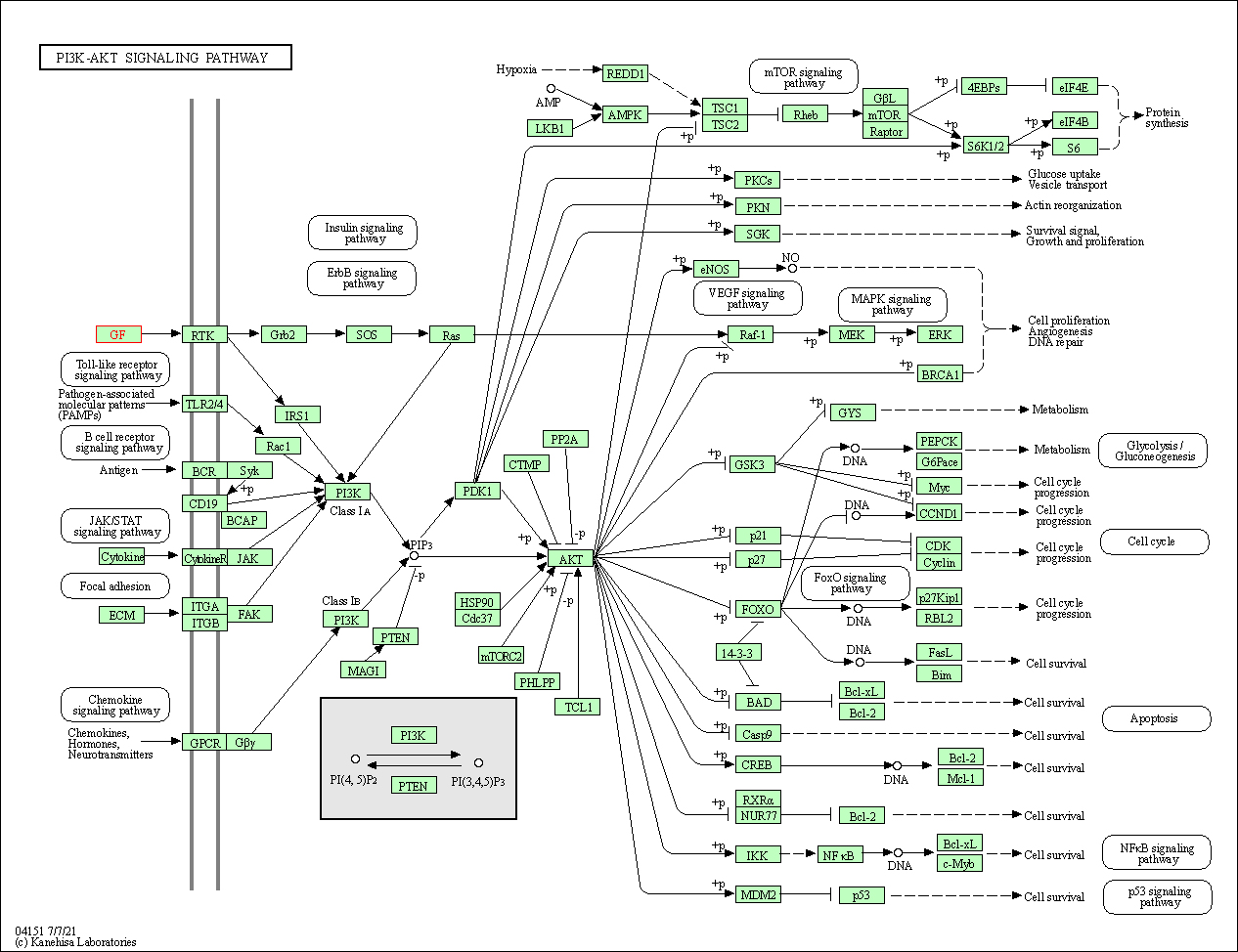
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
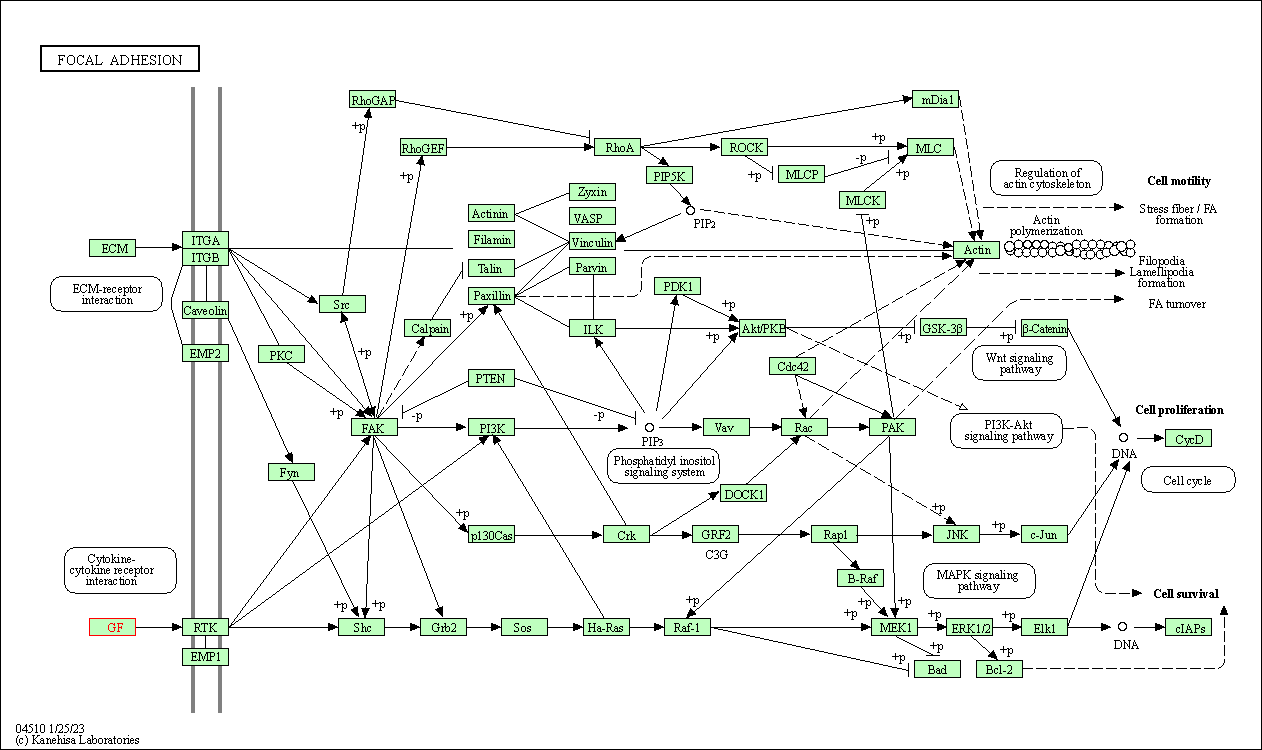
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 25 | Degree centrality | 2.69E-03 | Betweenness centrality | 2.88E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.49E-01 | Radiality | 1.43E+01 | Clustering coefficient | 2.07E-01 |
| Neighborhood connectivity | 5.25E+01 | Topological coefficient | 7.90E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Therapeutic angiogenesis using naked DNA expressing two isoforms of the hepatocyte growth factor in a porcine acute myocardial infarction model. Eur J Cardiothorac Surg. 2008 Oct;34(4):857-63. | |||||
| REF 2 | ClinicalTrials.gov (NCT02474667) A Multicenter, Prospective, Double-Blind, Randomized, Placebo-Controlled, Phase 3 Study of ANG-3777 (Formerly BB3) to Improve Graft Function and Reduce the Severity of Kidney Dysfunction or Delayed Graft Function Following Kidney Transplantation in Recipients of a Deceased Donor Kidney. U.S.National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT02563522) Safety and Efficacy Study of VM202 in the Treatment of Chronic Non-Healing Foot Ulcers. U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT02427464) Phase 3 Gene Therapy for Painful Diabetic Neuropathy. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT04267640) A Phase II Double-Blind, Randomized, Placebo-Controlled Study to Assess the Efficacy and Safety of AMG0001 to Improve Ulcer Healing and Perfusion in Subjects Who Have Peripheral Ischemic Ulcers of the Lower Extremity. U.S.National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT02318368) A Phase 2, Multicenter, Study of Ficlatuzumab Plus Erlotinib Versus Placebo Plus Erlotinib in Subjects Who Have Previously Untreated Metastatic, EGFR-mutated Non-small Cell Lung Cancer (NSCLC) and BDX004 Positive Label. U.S. National Institutes of Health. | |||||
| REF 7 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 8 | ClinicalTrials.gov (NCT01163071) A Phase 1 Trial of ABI-011 in Patients With Advanced Solid Tumors or Lymphomas. U.S. National Institutes of Health. | |||||
| REF 9 | ClinicalTrials.gov (NCT00831896) Phase I Study of TAK-701 in Adult Patients With Advanced Nonhematological Malignancies. U.S. National Institutes of Health. | |||||
| REF 10 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7986). | |||||
| REF 11 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800015060) | |||||
| REF 12 | Hepatocyte Growth Factor Mimetic ANG-3777 for Cardiac Surgery-Associated Acute Kidney Injury. Kidney Int Rep. 2020 Sep 25;5(12):2325-2332. | |||||
| REF 13 | Clinical pipeline report, company report or official report of ViroMed. | |||||
| REF 14 | Randomized, double-blind, placebo-controlled clinical trial of hepatocyte growth factor plasmid for critical limb ischemia. Gene Ther. 2010 Sep;17(9):1152-61. | |||||
| REF 15 | Targeting MET in cancer: rationale and progress.Nat Rev Cancer.2012 Jan 24;12(2):89-103. | |||||
| REF 16 | Clinical pipeline report, company report or official report of Amgen (2009). | |||||
| REF 17 | AMG 102, a fully human anti-hepatocyte growth factor/scatter factor neutralizing antibody, enhances the efficacy of temozolomide or docetaxel in U-87 MG cells and xenografts. Clin Cancer Res. 2007 Nov 15;13(22 Pt 1):6735-42. | |||||
| REF 18 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 19 | Engineering a fragment of Hepatocyte Growth Factor/Scatter Factor for tissue and organ regeneration | |||||
| REF 20 | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding. Chem Sci. 2015 Nov 1;6(11):6147-6157. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

