Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T91130
(Former ID: TTDR01159)
|
|||||
| Target Name |
LDL receptor related protein-1 (LRP-1)
|
|||||
| Synonyms |
Alpha-2-macroglobulin receptor; A2MR
Click to Show/Hide
|
|||||
| Gene Name |
LRP1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Required for early embryonic development. Involved in cellular lipid homeostasis. Involved in the plasma clearance of chylomicron remnants and activated LRPAP1 (alpha 2-macroglobulin), as well as the local metabolism of complexes between plasminogen activators and their endogenous inhibitors. May modulate cellular events, such as APP metabolism, kinase-dependent intracellular signaling, neuronal calcium signaling as well as neurotransmission. Acts as an alpha-2-macroglobulin receptor. Endocytic receptor involved in endocytosis and in phagocytosis of apoptotic cells.
Click to Show/Hide
|
|||||
| BioChemical Class |
Low density lipoprotein receptor
|
|||||
| UniProt ID | ||||||
| Sequence |
MLTPPLLLLLPLLSALVAAAIDAPKTCSPKQFACRDQITCISKGWRCDGERDCPDGSDEA
PEICPQSKAQRCQPNEHNCLGTELCVPMSRLCNGVQDCMDGSDEGPHCRELQGNCSRLGC QHHCVPTLDGPTCYCNSSFQLQADGKTCKDFDECSVYGTCSQLCTNTDGSFICGCVEGYL LQPDNRSCKAKNEPVDRPPVLLIANSQNILATYLSGAQVSTITPTSTRQTTAMDFSYANE TVCWVHVGDSAAQTQLKCARMPGLKGFVDEHTINISLSLHHVEQMAIDWLTGNFYFVDDI DDRIFVCNRNGDTCVTLLDLELYNPKGIALDPAMGKVFFTDYGQIPKVERCDMDGQNRTK LVDSKIVFPHGITLDLVSRLVYWADAYLDYIEVVDYEGKGRQTIIQGILIEHLYGLTVFE NYLYATNSDNANAQQKTSVIRVNRFNSTEYQVVTRVDKGGALHIYHQRRQPRVRSHACEN DQYGKPGGCSDICLLANSHKARTCRCRSGFSLGSDGKSCKKPEHELFLVYGKGRPGIIRG MDMGAKVPDEHMIPIENLMNPRALDFHAETGFIYFADTTSYLIGRQKIDGTERETILKDG IHNVEGVAVDWMGDNLYWTDDGPKKTISVARLEKAAQTRKTLIEGKMTHPRAIVVDPLNG WMYWTDWEEDPKDSRRGRLERAWMDGSHRDIFVTSKTVLWPNGLSLDIPAGRLYWVDAFY DRIETILLNGTDRKIVYEGPELNHAFGLCHHGNYLFWTEYRSGSVYRLERGVGGAPPTVT LLRSERPPIFEIRMYDAQQQQVGTNKCRVNNGGCSSLCLATPGSRQCACAEDQVLDADGV TCLANPSYVPPPQCQPGEFACANSRCIQERWKCDGDNDCLDNSDEAPALCHQHTCPSDRF KCENNRCIPNRWLCDGDNDCGNSEDESNATCSARTCPPNQFSCASGRCIPISWTCDLDDD CGDRSDESASCAYPTCFPLTQFTCNNGRCININWRCDNDNDCGDNSDEAGCSHSCSSTQF KCNSGRCIPEHWTCDGDNDCGDYSDETHANCTNQATRPPGGCHTDEFQCRLDGLCIPLRW RCDGDTDCMDSSDEKSCEGVTHVCDPSVKFGCKDSARCISKAWVCDGDNDCEDNSDEENC ESLACRPPSHPCANNTSVCLPPDKLCDGNDDCGDGSDEGELCDQCSLNNGGCSHNCSVAP GEGIVCSCPLGMELGPDNHTCQIQSYCAKHLKCSQKCDQNKFSVKCSCYEGWVLEPDGES CRSLDPFKPFIIFSNRHEIRRIDLHKGDYSVLVPGLRNTIALDFHLSQSALYWTDVVEDK IYRGKLLDNGALTSFEVVIQYGLATPEGLAVDWIAGNIYWVESNLDQIEVAKLDGTLRTT LLAGDIEHPRAIALDPRDGILFWTDWDASLPRIEAASMSGAGRRTVHRETGSGGWPNGLT VDYLEKRILWIDARSDAIYSARYDGSGHMEVLRGHEFLSHPFAVTLYGGEVYWTDWRTNT LAKANKWTGHNVTVVQRTNTQPFDLQVYHPSRQPMAPNPCEANGGQGPCSHLCLINYNRT VSCACPHLMKLHKDNTTCYEFKKFLLYARQMEIRGVDLDAPYYNYIISFTVPDIDNVTVL DYDAREQRVYWSDVRTQAIKRAFINGTGVETVVSADLPNAHGLAVDWVSRNLFWTSYDTN KKQINVARLDGSFKNAVVQGLEQPHGLVVHPLRGKLYWTDGDNISMANMDGSNRTLLFSG QKGPVGLAIDFPESKLYWISSGNHTINRCNLDGSGLEVIDAMRSQLGKATALAIMGDKLW WADQVSEKMGTCSKADGSGSVVLRNSTTLVMHMKVYDESIQLDHKGTNPCSVNNGDCSQL CLPTSETTRSCMCTAGYSLRSGQQACEGVGSFLLYSVHEGIRGIPLDPNDKSDALVPVSG TSLAVGIDFHAENDTIYWVDMGLSTISRAKRDQTWREDVVTNGIGRVEGIAVDWIAGNIY WTDQGFDVIEVARLNGSFRYVVISQGLDKPRAITVHPEKGYLFWTEWGQYPRIERSRLDG TERVVLVNVSISWPNGISVDYQDGKLYWCDARTDKIERIDLETGENREVVLSSNNMDMFS VSVFEDFIYWSDRTHANGSIKRGSKDNATDSVPLRTGIGVQLKDIKVFNRDRQKGTNVCA VANGGCQQLCLYRGRGQRACACAHGMLAEDGASCREYAGYLLYSERTILKSIHLSDERNL NAPVQPFEDPEHMKNVIALAFDYRAGTSPGTPNRIFFSDIHFGNIQQINDDGSRRITIVE NVGSVEGLAYHRGWDTLYWTSYTTSTITRHTVDQTRPGAFERETVITMSGDDHPRAFVLD ECQNLMFWTNWNEQHPSIMRAALSGANVLTLIEKDIRTPNGLAIDHRAEKLYFSDATLDK IERCEYDGSHRYVILKSEPVHPFGLAVYGEHIFWTDWVRRAVQRANKHVGSNMKLLRVDI PQQPMGIIAVANDTNSCELSPCRINNGGCQDLCLLTHQGHVNCSCRGGRILQDDLTCRAV NSSCRAQDEFECANGECINFSLTCDGVPHCKDKSDEKPSYCNSRRCKKTFRQCSNGRCVS NMLWCNGADDCGDGSDEIPCNKTACGVGEFRCRDGTCIGNSSRCNQFVDCEDASDEMNCS ATDCSSYFRLGVKGVLFQPCERTSLCYAPSWVCDGANDCGDYSDERDCPGVKRPRCPLNY FACPSGRCIPMSWTCDKEDDCEHGEDETHCNKFCSEAQFECQNHRCISKQWLCDGSDDCG DGSDEAAHCEGKTCGPSSFSCPGTHVCVPERWLCDGDKDCADGADESIAAGCLYNSTCDD REFMCQNRQCIPKHFVCDHDRDCADGSDESPECEYPTCGPSEFRCANGRCLSSRQWECDG ENDCHDQSDEAPKNPHCTSQEHKCNASSQFLCSSGRCVAEALLCNGQDDCGDSSDERGCH INECLSRKLSGCSQDCEDLKIGFKCRCRPGFRLKDDGRTCADVDECSTTFPCSQRCINTH GSYKCLCVEGYAPRGGDPHSCKAVTDEEPFLIFANRYYLRKLNLDGSNYTLLKQGLNNAV ALDFDYREQMIYWTDVTTQGSMIRRMHLNGSNVQVLHRTGLSNPDGLAVDWVGGNLYWCD KGRDTIEVSKLNGAYRTVLVSSGLREPRALVVDVQNGYLYWTDWGDHSLIGRIGMDGSSR SVIVDTKITWPNGLTLDYVTERIYWADAREDYIEFASLDGSNRHVVLSQDIPHIFALTLF EDYVYWTDWETKSINRAHKTTGTNKTLLISTLHRPMDLHVFHALRQPDVPNHPCKVNNGG CSNLCLLSPGGGHKCACPTNFYLGSDGRTCVSNCTASQFVCKNDKCIPFWWKCDTEDDCG DHSDEPPDCPEFKCRPGQFQCSTGICTNPAFICDGDNDCQDNSDEANCDIHVCLPSQFKC TNTNRCIPGIFRCNGQDNCGDGEDERDCPEVTCAPNQFQCSITKRCIPRVWVCDRDNDCV DGSDEPANCTQMTCGVDEFRCKDSGRCIPARWKCDGEDDCGDGSDEPKEECDERTCEPYQ FRCKNNRCVPGRWQCDYDNDCGDNSDEESCTPRPCSESEFSCANGRCIAGRWKCDGDHDC ADGSDEKDCTPRCDMDQFQCKSGHCIPLRWRCDADADCMDGSDEEACGTGVRTCPLDEFQ CNNTLCKPLAWKCDGEDDCGDNSDENPEECARFVCPPNRPFRCKNDRVCLWIGRQCDGTD NCGDGTDEEDCEPPTAHTTHCKDKKEFLCRNQRCLSSSLRCNMFDDCGDGSDEEDCSIDP KLTSCATNASICGDEARCVRTEKAAYCACRSGFHTVPGQPGCQDINECLRFGTCSQLCNN TKGGHLCSCARNFMKTHNTCKAEGSEYQVLYIADDNEIRSLFPGHPHSAYEQAFQGDESV RIDAMDVHVKAGRVYWTNWHTGTISYRSLPPAAPPTTSNRHRRQIDRGVTHLNISGLKMP RGIAIDWVAGNVYWTDSGRDVIEVAQMKGENRKTLISGMIDEPHAIVVDPLRGTMYWSDW GNHPKIETAAMDGTLRETLVQDNIQWPTGLAVDYHNERLYWADAKLSVIGSIRLNGTDPI VAADSKRGLSHPFSIDVFEDYIYGVTYINNRVFKIHKFGHSPLVNLTGGLSHASDVVLYH QHKQPEVTNPCDRKKCEWLCLLSPSGPVCTCPNGKRLDNGTCVPVPSPTPPPDAPRPGTC NLQCFNGGSCFLNARRQPKCRCQPRYTGDKCELDQCWEHCRNGGTCAASPSGMPTCRCPT GFTGPKCTQQVCAGYCANNSTCTVNQGNQPQCRCLPGFLGDRCQYRQCSGYCENFGTCQM AADGSRQCRCTAYFEGSRCEVNKCSRCLEGACVVNKQSGDVTCNCTDGRVAPSCLTCVGH CSNGGSCTMNSKMMPECQCPPHMTGPRCEEHVFSQQQPGHIASILIPLLLLLLLVLVAGV VFWYKRRVQGAKGFQHQRMTNGAMNVEIGNPTYKMYEGGEPDDVGGLLDADFALDPDKPT NFTNPVYATLYMGGHGSRHSLASTDEKRELLGRGPEDEIGDPLA Click to Show/Hide
|
|||||
| HIT2.0 ID | T79UHV | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
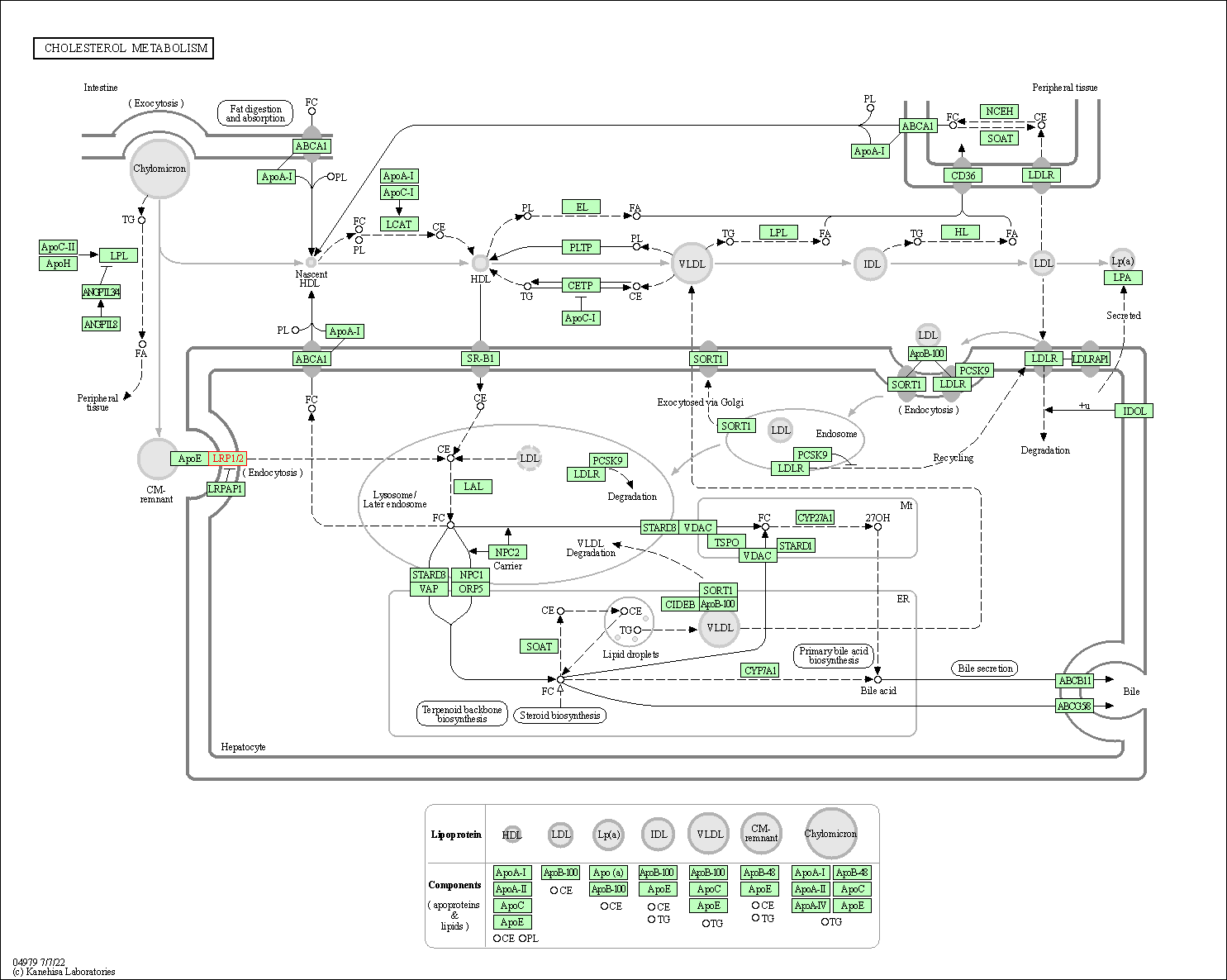
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cholesterol metabolism | hsa04979 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 23 | Degree centrality | 2.47E-03 | Betweenness centrality | 3.14E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.43E-01 | Radiality | 1.43E+01 | Clustering coefficient | 5.93E-02 |
| Neighborhood connectivity | 3.04E+01 | Topological coefficient | 6.03E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Expression of LRP1 in retinal pigment epithelial cells and its regulation by growth factors. Invest Ophthalmol Vis Sci. 2004 Jun;45(6):2033-8. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

