Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T95736
(Former ID: TTDI03381)
|
|||||
| Target Name |
Serine/threonine-protein kinase NIK (MAP3K14)
|
|||||
| Synonyms |
NIK; NF-kappa-beta-inducing kinase; Mitogen-activated protein kinase kinase kinase 14; HsNIK
Click to Show/Hide
|
|||||
| Gene Name |
MAP3K14
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Promotes proteolytic processing of NFKB2/P100, which leads to activation of NF-kappa-B via the non-canonical pathway. Could act in a receptor-selective manner. Lymphotoxin beta-activated kinase which seems to be exclusively involved in the activation of NF-kappa-B and its transcriptional activity.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.25
|
|||||
| Sequence |
MAVMEMACPGAPGSAVGQQKELPKAKEKTPPLGKKQSSVYKLEAVEKSPVFCGKWEILND
VITKGTAKEGSEAGPAAISIIAQAECENSQEFSPTFSERIFIAGSKQYSQSESLDQIPNN VAHATEGKMARVCWKGKRRSKARKKRKKKSSKSLAHAGVALAKPLPRTPEQESCTIPVQE DESPLGAPYVRNTPQFTKPLKEPGLGQLCFKQLGEGLRPALPRSELHKLISPLQCLNHVW KLHHPQDGGPLPLPTHPFPYSRLPHPFPFHPLQPWKPHPLESFLGKLACVDSQKPLPDPH LSKLACVDSPKPLPGPHLEPSCLSRGAHEKFSVEEYLVHALQGSVSSGQAHSLTSLAKTW AARGSRSREPSPKTEDNEGVLLTEKLKPVDYEYREEVHWATHQLRLGRGSFGEVHRMEDK QTGFQCAVKKVRLEVFRAEELMACAGLTSPRIVPLYGAVREGPWVNIFMELLEGGSLGQL VKEQGCLPEDRALYYLGQALEGLEYLHSRRILHGDVKADNVLLSSDGSHAALCDFGHAVC LQPDGLGKSLLTGDYIPGTETHMAPEVVLGRSCDAKVDVWSSCCMMLHMLNGCHPWTQFF RGPLCLKIASEPPPVREIPPSCAPLTAQAIQEGLRKEPIHRVSAAELGGKVNRALQQVGG LKSPWRGEYKEPRHPPPNQANYHQTLHAQPRELSPRAPGPRPAEETTGRAPKLQPPLPPE PPEPNKSPPLTLSKEESGMWEPLPLSSLEPAPARNPSSPERKATVPEQELQQLEIELFLN SLSQPFSLEEQEQILSCLSIDSLSLSDDSEKNPSKASQSSRDTLSSGVHSWSSQAEARSS SWNMVLARGRPTDTPSYFNGVKVQIQSLNGEHLHIREFHRVKVGDIATGISSQIPAAAFS LVTKDGQPVRYDMEVPDSGIDLQCTLAPDGSFAWSWRVKHGQLENRP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: PMID23374866C31 | Ligand Info | |||||
| Structure Description | Crystal Structure of NIK with compound 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol (13V) | PDB:4IDV | ||||
| Method | X-ray diffraction | Resolution | 2.90 Å | Mutation | No | [1] |
| PDB Sequence |
FSVEEYLVHA
340 LQGSVSSGQA350 HSLTSLAKTW360 AARTEDNEGV380 LLTEKLKPVD390 YEYREEVHWA 400 THQLRLGRGS410 FGEVHRMEDK420 QTGFQCAVKK430 VRLEVFRAEE440 LMACAGLTSP 450 RIVPLYGAVR460 EGPWVNIFME470 LLEGGSLGQL480 VKEQGCLPED490 RALYYLGQAL 500 EGLEYLHSRR510 ILHGDVKADN520 VLLSSDGSHA530 ALCDFGHAVC540 LQPDGLGKSL 550 LTGDYIPGTE560 THMAPEVVLG570 RSCDAKVDVW580 SSCCMMLHML590 NGCHPWTQFF 600 RGPLCLKIAS610 EPPPVREIPP620 SCAPLTAQAI630 QEGLRKEPIH640 RVSAAELGGK 650 VNRALQQVGG660 LKSPWRGEYK670 EPRHP
|
|||||
|
|
ARG408
3.487
GLY409
3.634
GLU413
4.523
VAL414
3.470
ALA427
3.930
LYS429
3.898
GLU440
2.624
CYS444
4.196
VAL453
3.878
ILE467
3.595
MET469
3.479
GLU470
2.614
|
|||||
| Ligand Name: 11-Bromo-5,6,7,8-Tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-B]indol-2-Amine | Ligand Info | |||||
| Structure Description | Crystal Structure of NIK with 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine (T28) | PDB:4IDT | ||||
| Method | X-ray diffraction | Resolution | 2.40 Å | Mutation | No | [1] |
| PDB Sequence |
VEEYLVHALQ
342 GSVSSGQAHS352 LTSLAKTWAA362 RGSRSREPSP372 KTEDNEGVLL382 TEKLKPVDYE 392 YREEVHWATH402 QLRLGRGSFG412 EVHRMEDKQT422 GFQCAVKKVR432 LEVFRAEELM 442 ACAGLTSPRI452 VPLYGAVREG462 PWVNIFMELL472 EGGSLGQLVK482 EQGCLPEDRA 492 LYYLGQALEG502 LEYLHSRRIL512 HGDVKADNVL522 LSSDGSHAAL532 CDFGHAVCLQ 542 TGDYIPGTET561 HMAPEVVLGR571 SCDAKVDVWS581 SCCMMLHMLN591 GCHPWTQFFR 601 GPLCLKIASE611 PPPVREIPPS621 CAPLTAQAIQ631 EGLRKEPIHR641 VSAAELGGKV 651 NRALQQVGGL661 KSPWRGEYKE671 PRHP
|
|||||
|
|
LEU406
2.129
GLY407
2.845
ARG408
2.923
GLY409
2.582
SER410
4.024
VAL414
2.687
ALA427
2.562
LYS429
2.969
VAL453
4.063
MET469
2.492
GLU470
2.134
LEU471
2.447
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|


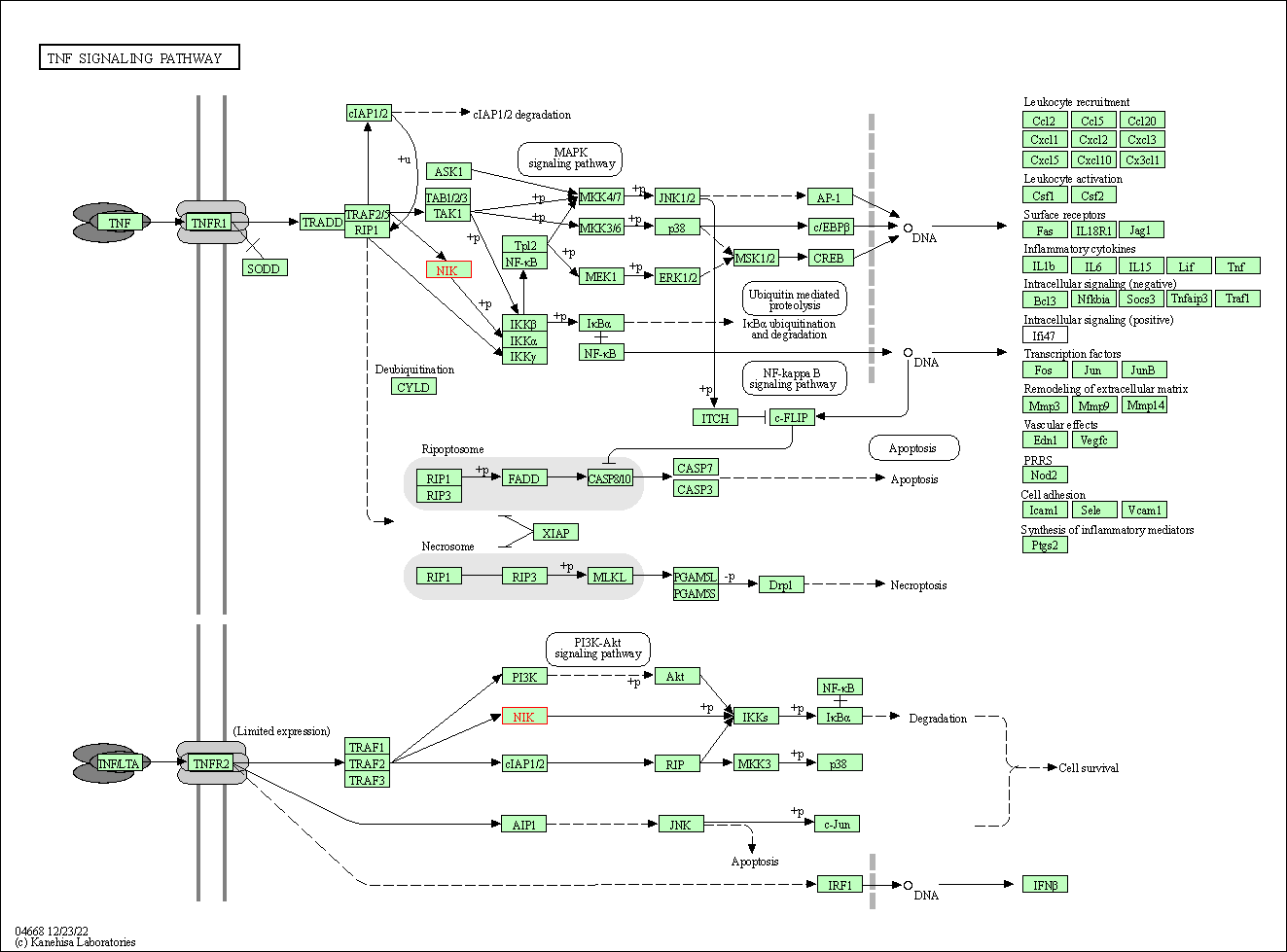
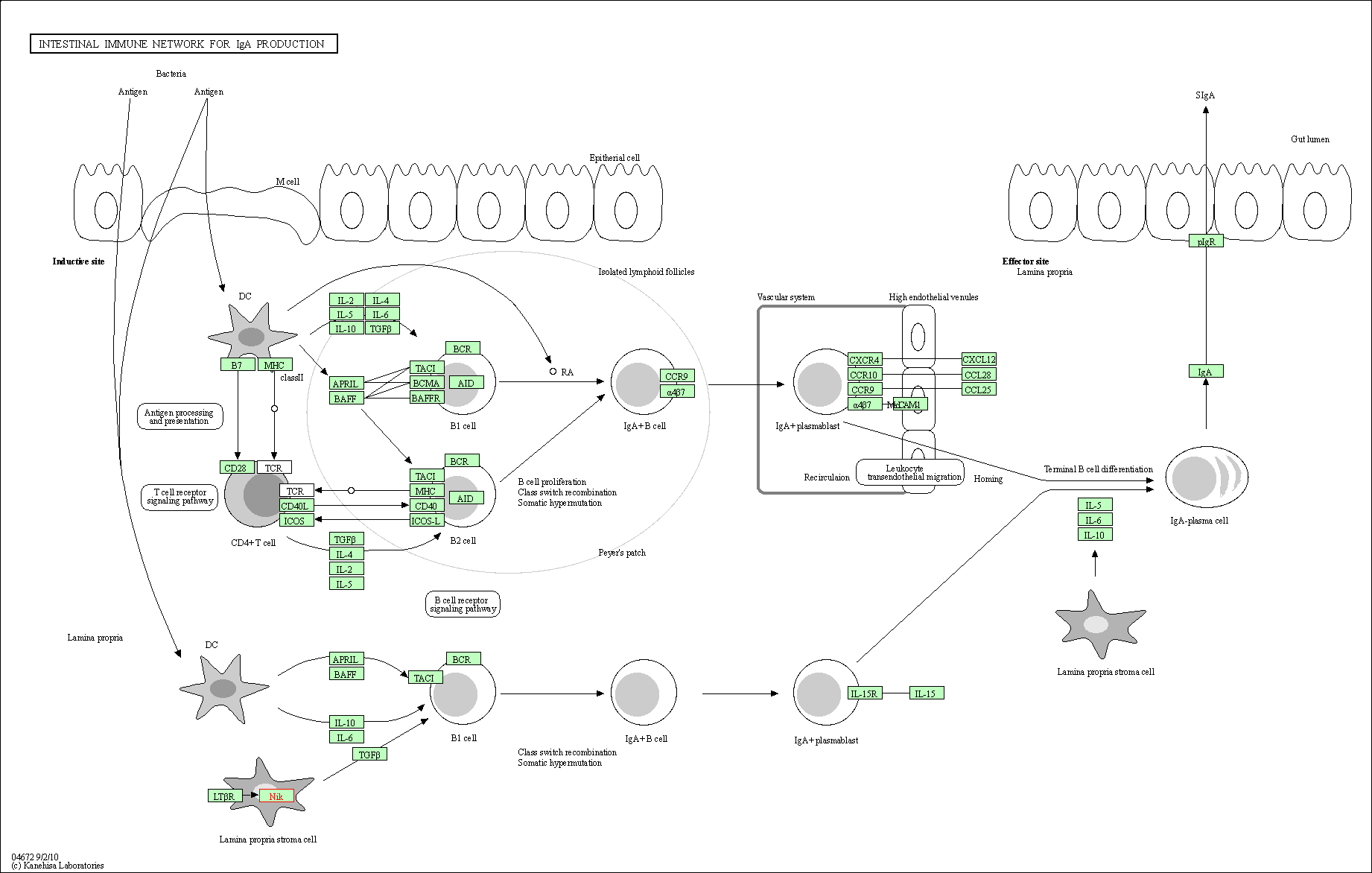
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
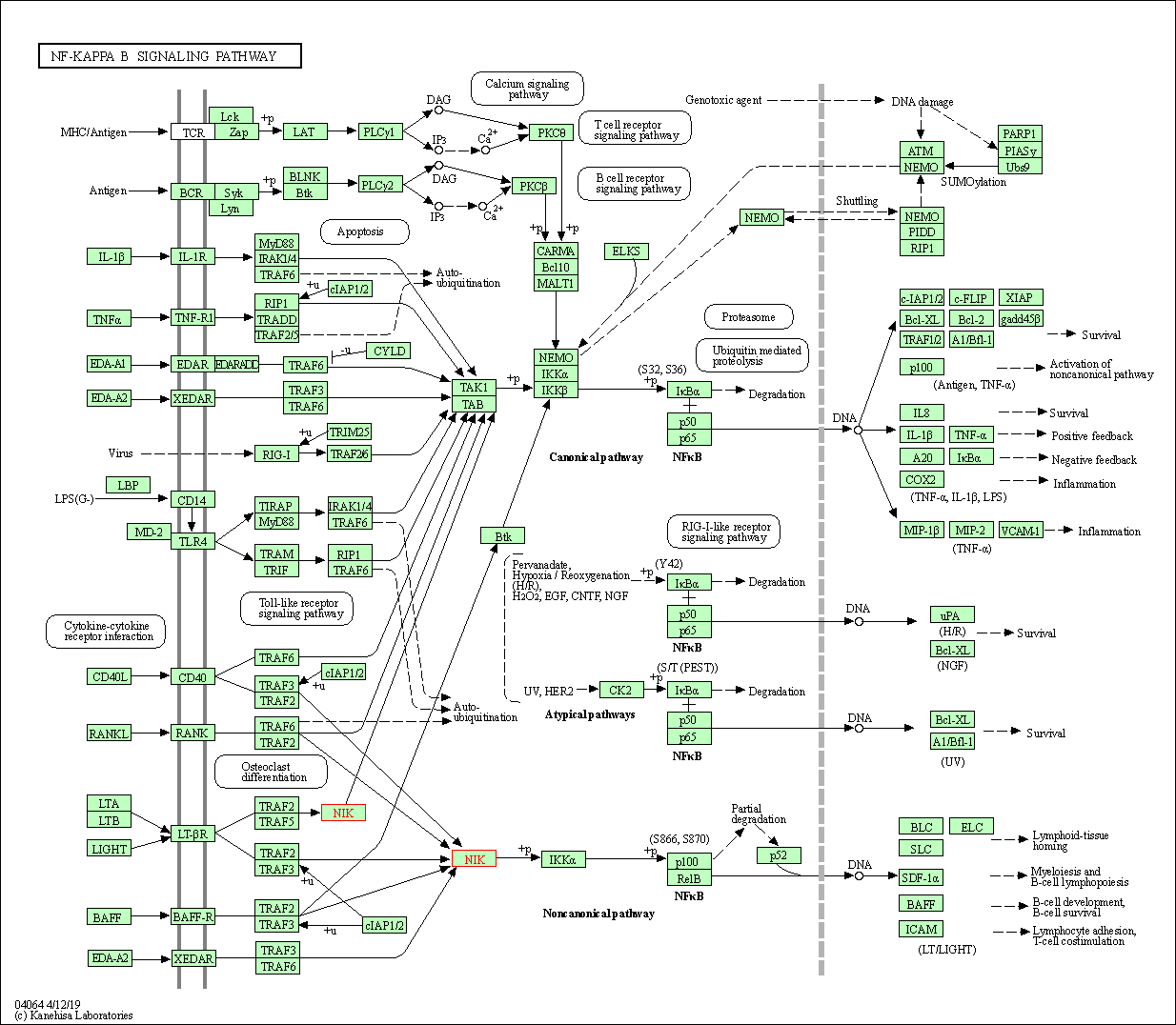
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
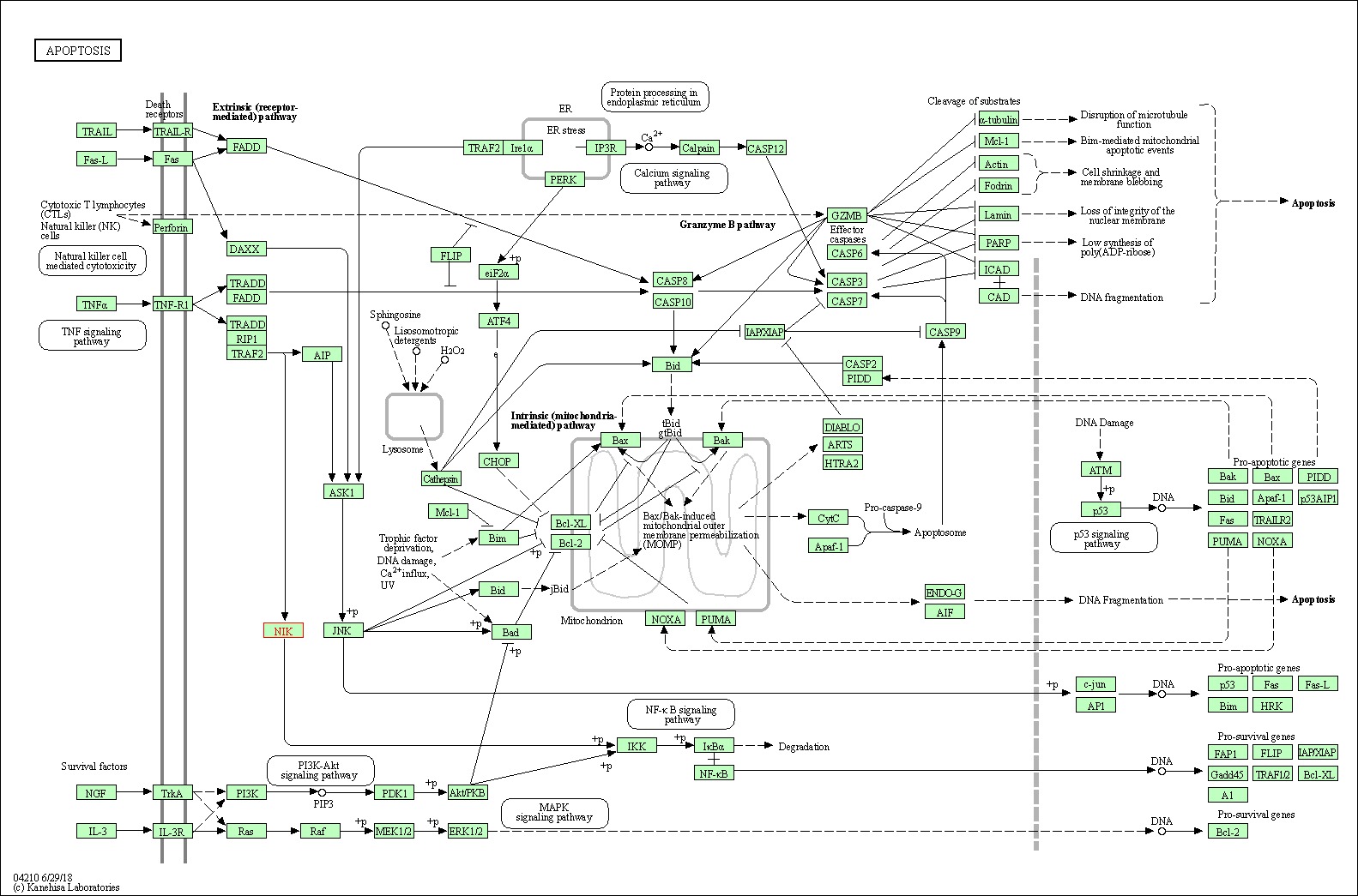
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Osteoclast differentiation | hsa04380 | Affiliated Target |

|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
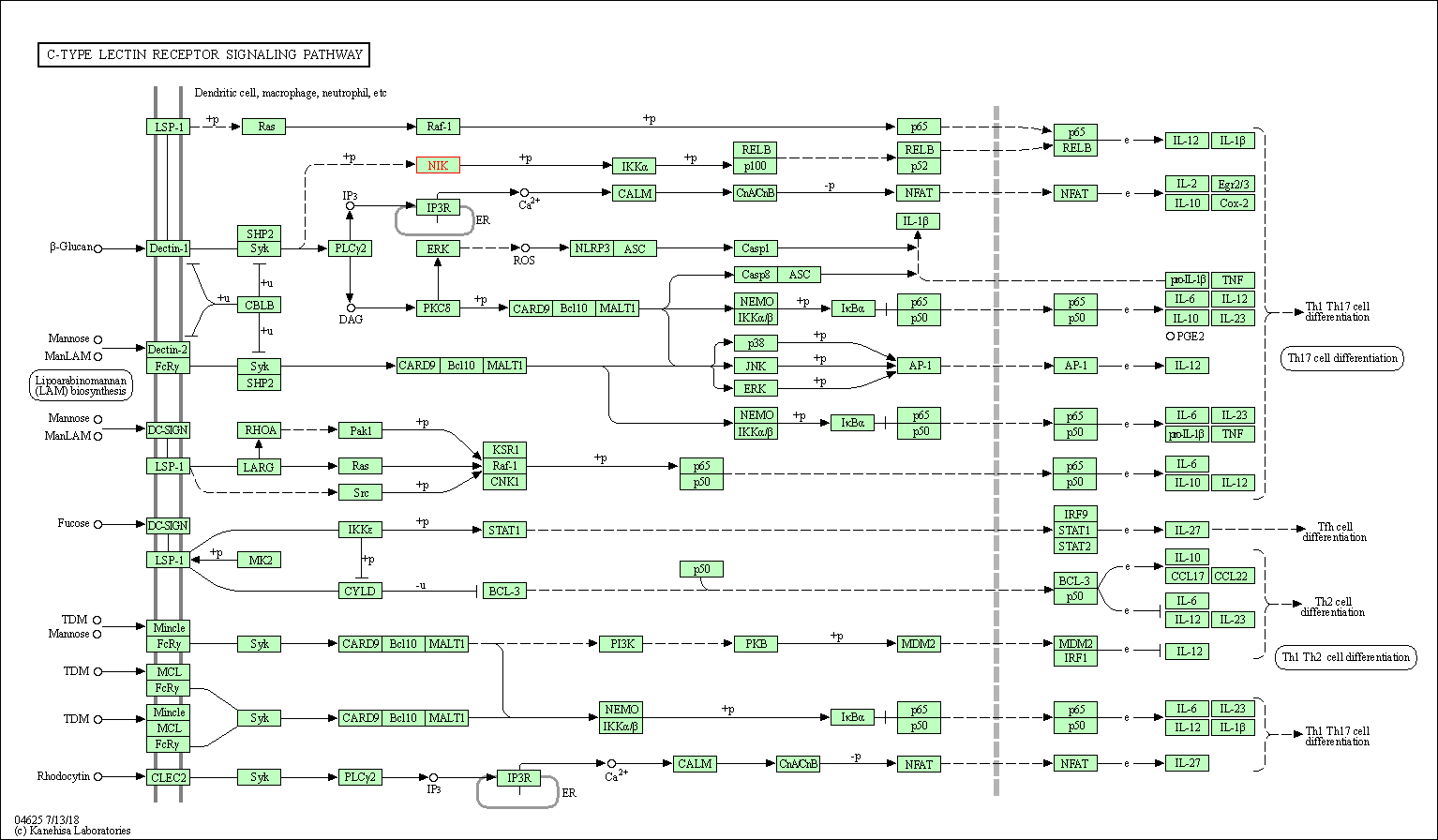
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
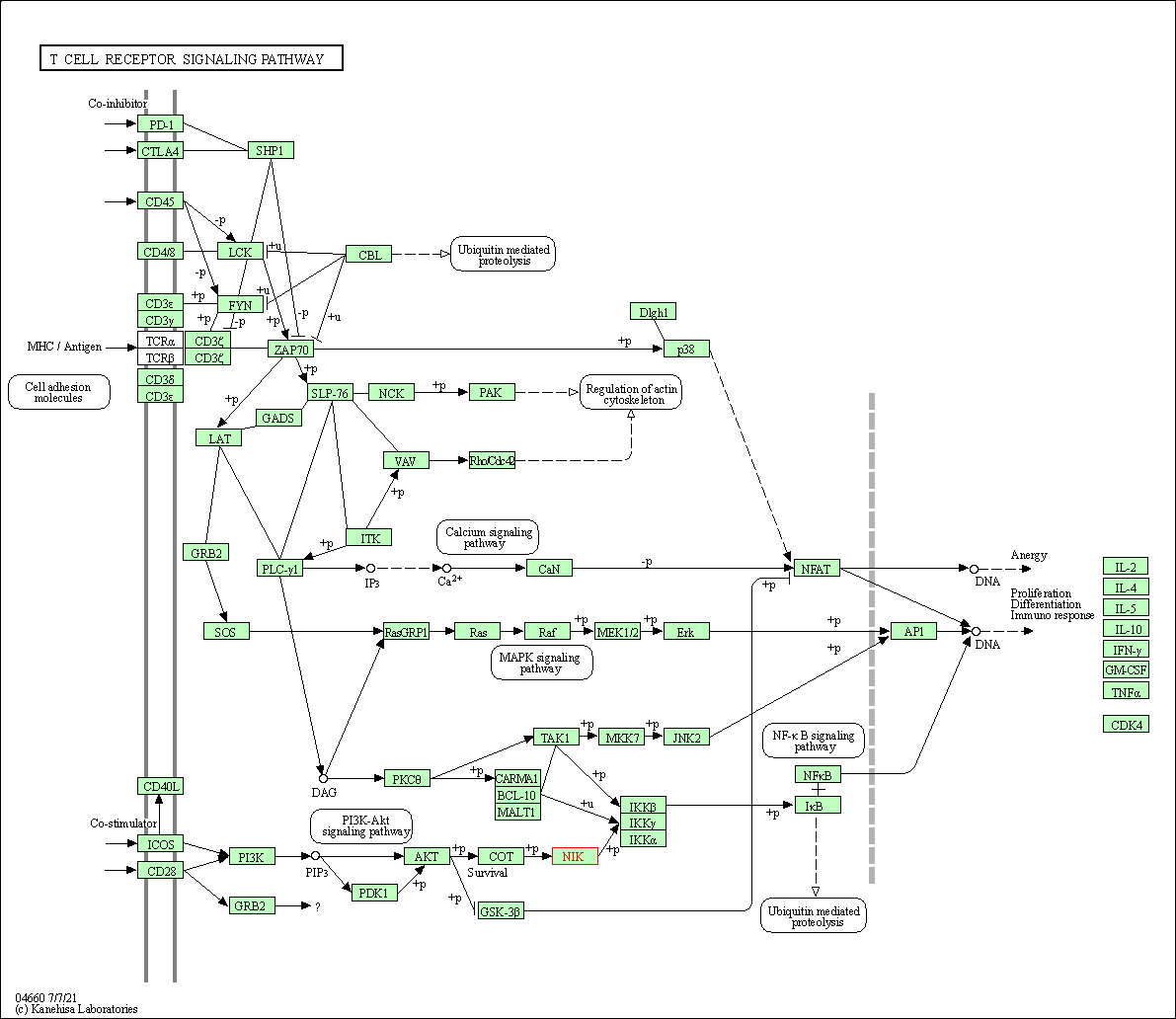
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Intestinal immune network for IgA production | hsa04672 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 14 | Degree centrality | 1.50E-03 | Betweenness centrality | 2.04E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.22E-01 | Radiality | 1.39E+01 | Clustering coefficient | 4.62E-01 |
| Neighborhood connectivity | 4.04E+01 | Topological coefficient | 1.91E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Inhibiting NF- B-inducing kinase (NIK): discovery, structure-based design, synthesis, structure-activity relationship, and co-crystal structures. Bioorg Med Chem Lett. 2013 Mar 1;23(5):1238-44. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

