Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T96623
|
|||||
| Target Name |
Heat shock protein 90 beta (HSP90B)
|
|||||
| Synonyms |
Heat shock 84 kDa; HSP84; HSP 84
Click to Show/Hide
|
|||||
| Gene Name |
HSP90AB1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 7 Target-related Diseases | + | ||||
| 1 | Lung cancer [ICD-11: 2C25] | |||||
| 2 | Breast cancer [ICD-11: 2C60-2C6Y] | |||||
| 3 | Malignant haematopoietic neoplasm [ICD-11: 2B33] | |||||
| 4 | Mature B-cell leukaemia [ICD-11: 2A82] | |||||
| 5 | Myeloproliferative neoplasm [ICD-11: 2A20] | |||||
| 6 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| 7 | Stomach cancer [ICD-11: 2B72] | |||||
| Function |
Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function (PubMed:16478993, PubMed:19696785). Engages with a range of client protein classes via its interaction with various co-chaperone proteins or complexes, that act as adapters, simultaneously able to interact with the specific client and the central chaperone itself. Recruitment of ATP and co-chaperone followed by client protein forms a functional chaperone. After the completion of the chaperoning process, properly folded client protein and co-chaperone leave HSP90 in an ADP-bound partially open conformation and finally, ADP is released from HSP90 which acquires an open conformation for the next cycle (PubMed:27295069, PubMed:26991466). Apart from its chaperone activity, it also plays a role in the regulation of the transcription machinery. HSP90 and its co-chaperones modulate transcription at least at three different levels. In the first place, they alter the steady-state levels of certain transcription factors in response to various physiological cues. Second, they modulate the activity of certain epigenetic modifiers, such as histone deacetylases or DNA methyl transferases, and thereby respond to the change in the environment. Third, they participate in the eviction of histones from the promoter region of certain genes and thereby turn on gene expression (PubMed:25973397). Antagonizes STUB1-mediated inhibition of TGF-beta signaling via inhibition of STUB1-mediated SMAD3 ubiquitination and degradation (PubMed:24613385). Promotes cell differentiation by chaperoning BIRC2 and thereby protecting from auto-ubiquitination and degradation by the proteasomal machinery (PubMed:18239673). Main chaperone that is involved in the phosphorylation/activation of the STAT1 by chaperoning both JAK2 and PRKCE under heat shock and in turn, activates its own transcription (PubMed:20353823).
Click to Show/Hide
|
|||||
| BioChemical Class |
Heat shock protein
|
|||||
| UniProt ID | ||||||
| Sequence |
MPEEVHHGEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLT
DPSKLDSGKELKIDIIPNPQERTLTLVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAG ADISMIGQFGVGFYSAYLVAEKVVVITKHNDDEQYAWESSAGGSFTVRADHGEPIGRGTK VILHLKEDQTEYLEERRVKEVVKKHSQFIGYPITLYLEKEREKEISDDEAEEEKGEKEEE DKDDEEKPKIEDVGSDEEDDSGKDKKKKTKKIKEKYIDQEELNKTKPIWTRNPDDITQEE YGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYVRRV FIMDSCDELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLELFSELA EDKENYKKFYEAFSKNLKLGIHEDSTNRRRLSELLRYHTSQSGDEMTSLSEYVSRMKETQ KSIYYITGESKEQVANSAFVERVRKRGFEVVYMTEPIDEYCVQQLKEFDGKSLVSVTKEG LELPEDEEEKKKMEESKAKFENLCKLMKEILDKKVEKVTISNRLVSSPCCIVTSTYGWTA NMERIMKAQALRDNSTMGYMMAKKHLEINPDHPIVETLRQKAEADKNDKAVKDLVVLLFE TALLSSGFSLEDPQTHSNRIYRMIKLGLGIDEDEVAAEEPNAAVPDEIPPLEGDEDASRM EEVD Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | SNX-5422 | Drug Info | Phase 1/2 | Haematological malignancy | [1] | |
| 2 | PU-H71 | Drug Info | Phase 1 | Solid tumour/cancer | [2] | |
| 3 | TAS-116 | Drug Info | Phase 1 | Solid tumour/cancer | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 3 Inhibitor drugs | + | ||||
| 1 | SNX-5422 | Drug Info | [1] | |||
| 2 | PU-H71 | Drug Info | [1] | |||
| 3 | TAS-116 | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | PDB:7Z38 | ||||
| Method | Electron microscopy | Resolution | 3.16 Å | Mutation | No | [3] |
| PDB Sequence |
EEVETFAFQA
19 EIAQLMSLII29 NTFYSNKEIF39 LRELISNASD49 ALDKIRYESL59 TDPSKLDSGK 69 ELKIDIIPNP79 QERTLTLVDT89 GIGMTKADLI99 NNLGTIAKSG109 TKAFMEALQA 119 GADISMIGQF129 GVGFYSAYLV139 AEKVVVITKH149 NDDEQYAWES159 SAGGSFTVRA 169 DHGEPIGRGT179 KVILHLKEDQ189 TEYLEERRVK199 EVVKKHSQFI209 GYPITLYLEK 219 EREKKKIKEK275 YIDQEELNKT285 KPIWTRNPDD295 ITQEEYGEFY305 KSLTNDWEDH 315 LAVKHFSVEG325 QLEFRALLFI335 PRRAPFDLFE345 NKKKKNNIKL355 YVRRVFIMDS 365 CDELIPEYLN375 FIRGVVDSED385 LPLNISREML395 QQSKILKVIR405 KNIVKKCLEL 415 FSELAEDKEN425 YKKFYEAFSK435 NLKLGIHEDS445 TNRRRLSELL455 RYHTSQSGDE 465 MTSLSEYVSR475 MKETQKSIYY485 ITGESKEQVA495 NSAFVERVRK505 RGFEVVYMTE 515 PIDEYCVQQL525 KEFDGKSLVS535 VTKEGLELPE545 DEEEKKKMEE555 SKAKFENLCK 565 LMKEILDKKV575 EKVTISNRLV585 SSPCCIVTST595 YGWTANMERI605 MKAQALRDNS 615 TMGYMMAKKH625 LEINPDHPIV635 ETLRQKAEAD645 KNDKAVKDLV655 VLLFETALLS 665 SGFSLEDPQT675 HSNRIYRMIK685 LGLGID
|
|||||
|
|
TYR33
4.837
GLU42
3.906
ASN46
2.626
ALA47
4.098
ASP49
3.965
ALA50
3.117
LYS53
3.986
ASP88
2.663
ILE91
4.212
GLY92
3.957
MET93
3.646
ASN101
3.326
LEU102
3.558
LYS107
3.967
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: PU3 | Ligand Info | |||||
| Structure Description | Human Hsp90-beta with PU3 (9-Butyl-8(3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine) | PDB:1UYM | ||||
| Method | X-ray diffraction | Resolution | 2.45 Å | Mutation | No | [4] |
| PDB Sequence |
EVETFAFQAE
25 IAQLMSLIIN35 TFYSNKEIFL45 RELISNASDA55 LDKIRYESLT65 DPSKLDSGKE 75 LKIDIIPNPQ85 ERTLTLVDTG95 IGMTKADLIN105 NLGTIAKSGT115 KAFMEALQAG 125 ADISMIGQFG135 VGFYSAYLVA145 EKVVVITKHN155 DDEQYAWESS165 AGGSFTVRAD 175 HGEPIGRGTK185 VILHLKEDQT195 EYLEERRVKE205 VVKKHSQFIG215 YPITLYLEKE 225 R
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
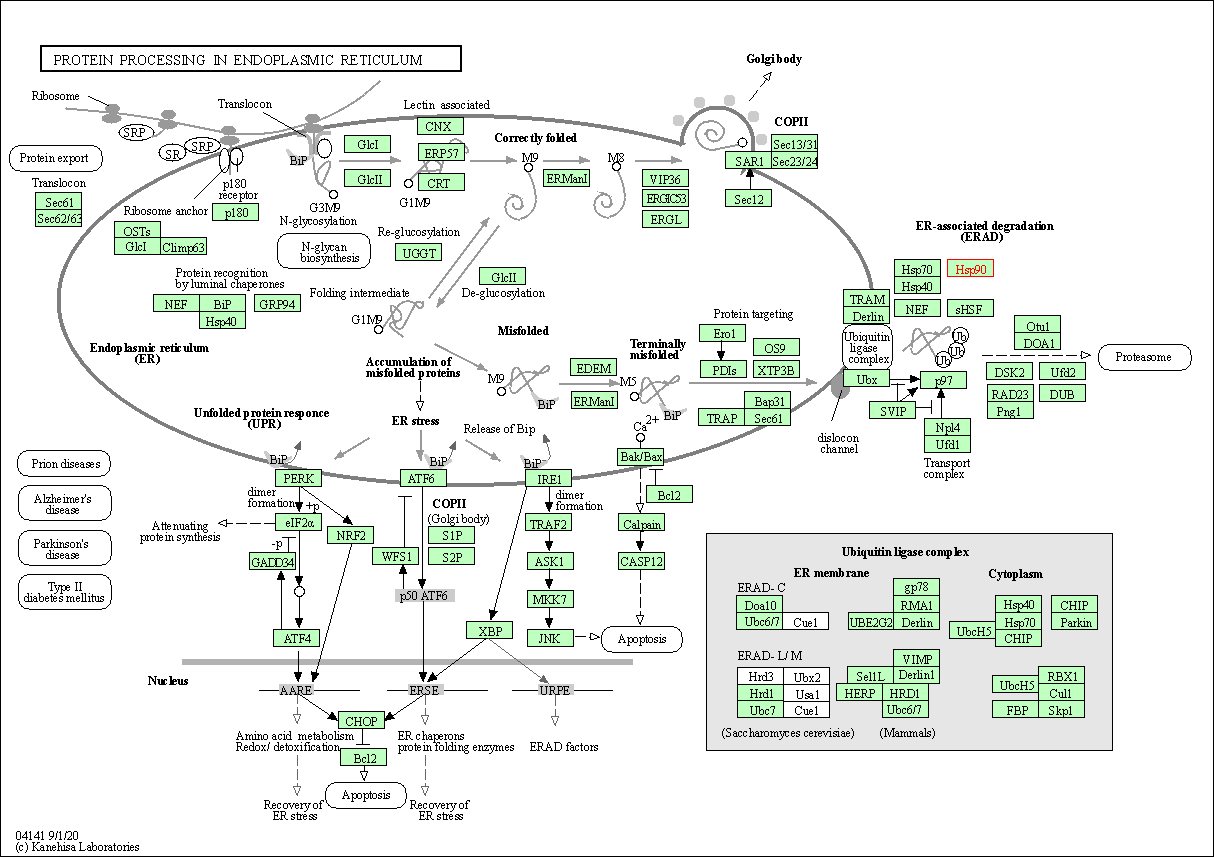
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |

|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
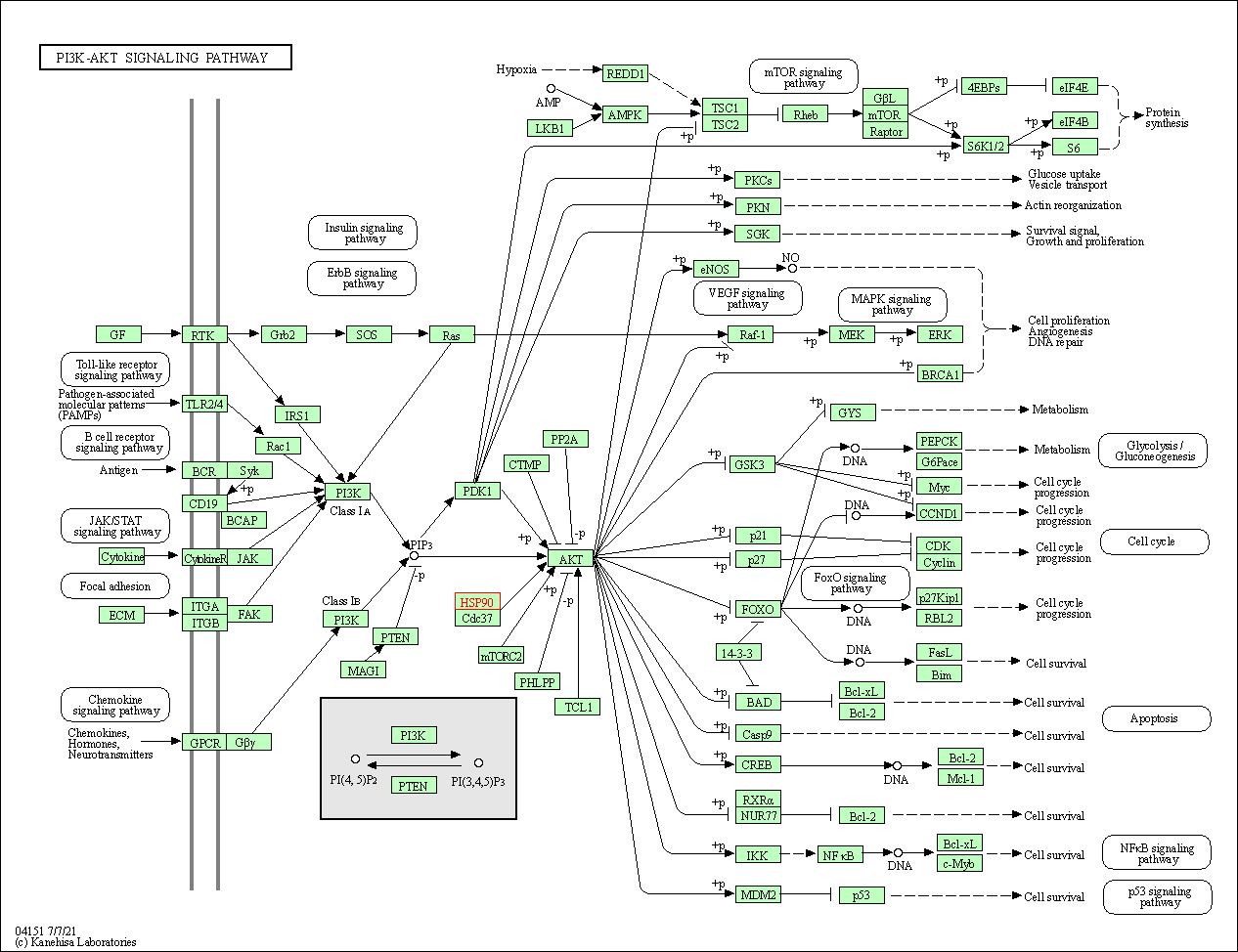
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
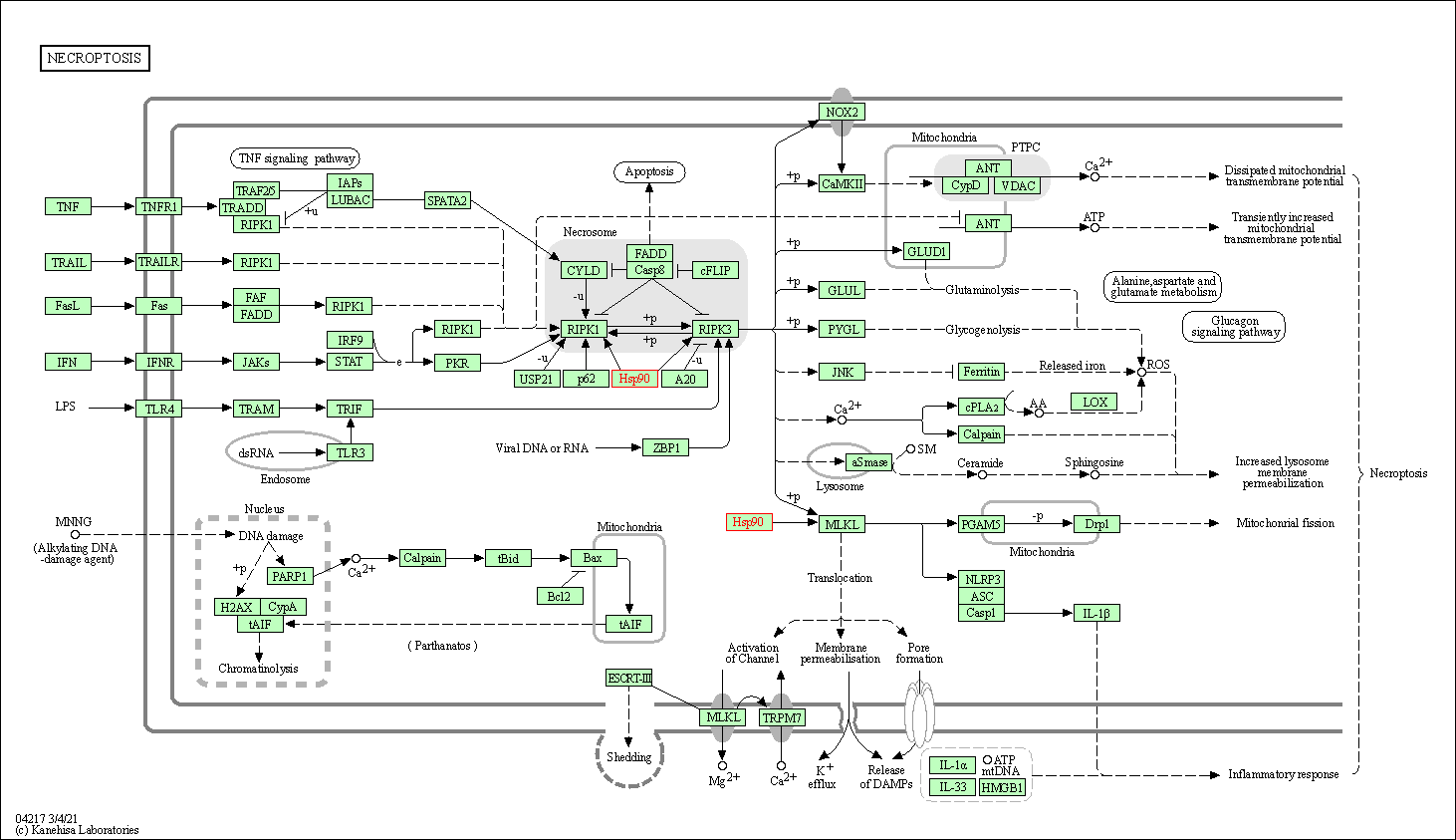
| Necroptosis | hsa04217 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
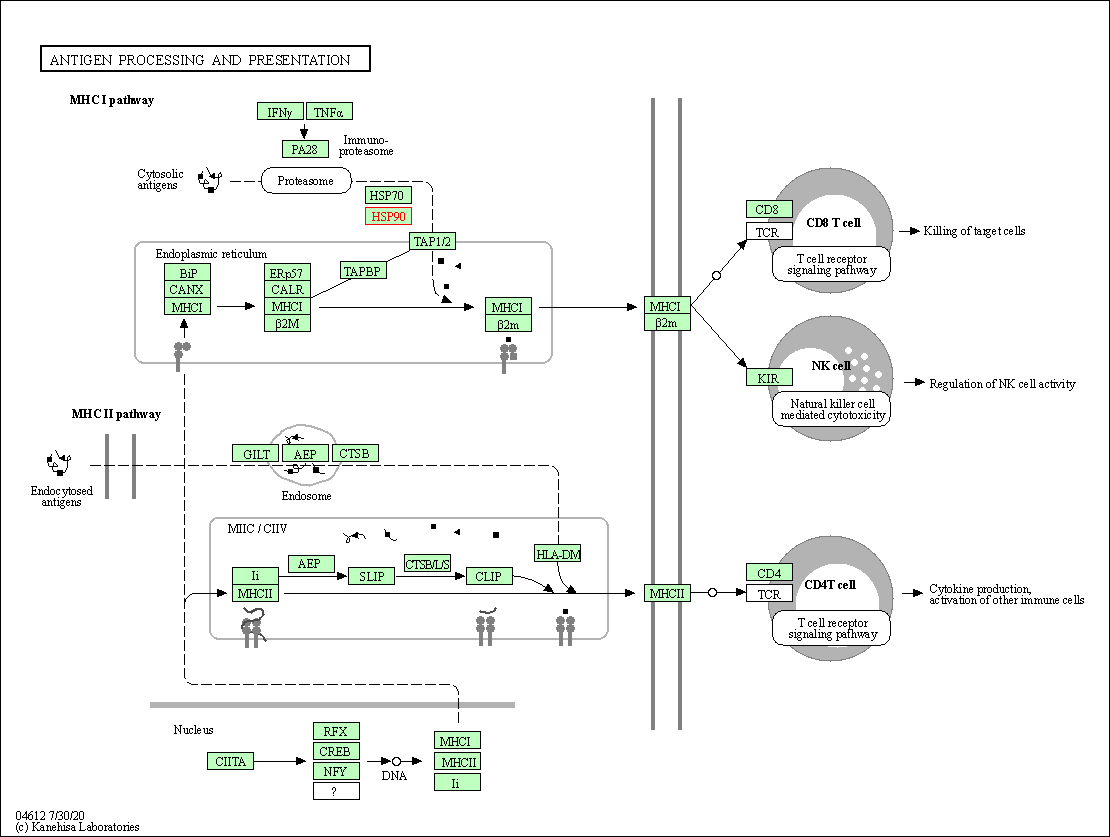
| Antigen processing and presentation | hsa04612 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Th17 cell differentiation | hsa04659 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 27 | Degree centrality | 2.90E-03 | Betweenness centrality | 1.18E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.57E-01 | Radiality | 1.44E+01 | Clustering coefficient | 2.11E-01 |
| Neighborhood connectivity | 4.37E+01 | Topological coefficient | 6.48E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 12 KEGG Pathways | + | ||||
| 1 | Protein processing in endoplasmic reticulum | |||||
| 2 | PI3K-Akt signaling pathway | |||||
| 3 | Necroptosis | |||||
| 4 | Antigen processing and presentation | |||||
| 5 | NOD-like receptor signaling pathway | |||||
| 6 | IL-17 signaling pathway | |||||
| 7 | Th17 cell differentiation | |||||
| 8 | Progesterone-mediated oocyte maturation | |||||
| 9 | Estrogen signaling pathway | |||||
| 10 | Pathways in cancer | |||||
| 11 | Prostate cancer | |||||
| 12 | Fluid shear stress and atherosclerosis | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013 Jan;41(Database issue):D991-5. | |||||
| REF 3 | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation. Mol Cell. 2022 Sep 15;82(18):3438-3452.e8. | |||||
| REF 4 | Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chem Biol. 2004 Jun;11(6):775-85. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

