| Drug General Information |
| Drug ID |
D06EWG
|
| Former ID |
DNC003012
|
| Drug Name |
Formic Acid
|
| Drug Type |
Small molecular drug
|
| Indication |
Discovery agent
|
Investigative |
[1]
|
|---|
| Structure |
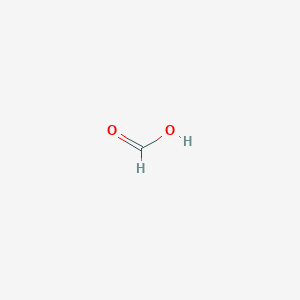
|
Download
2D MOL
3D MOL
|
| Formula |
CHO2-
|
| InChI |
InChI=1S/CH2O2/c2-1-3/h1H,(H,2,3)/p-1
|
| InChIKey |
BDAGIHXWWSANSR-UHFFFAOYSA-M
|
| CAS Number |
CAS 64-18-6
|
| PubChem Compound ID |
|
| PubChem Substance ID |
2203, 2712, 841549, 4668969, 8143755, 8150550, 10533692, 14709153, 24438659, 50073763, 50817872, 57320099, 92729992, 92741270, 103036860, 104042951, 104294385, 126406823, 126687287, 127497873, 135269870, 137002191, 142323754, 160644736, 160644737, 160644738, 163218836, 178101261, 223444138, 223444139, 223749098
|
| Target and Pathway |
| Target(s) |
3-phosphoshikimate1-carboxyvinyltransferase |
Target Info |
Inhibitor |
[2]
|
|---|
| Mannose-6-phosphate isomerase |
Target Info |
Inhibitor |
[2]
|
| Nitric-oxide synthase, brain |
Target Info |
Inhibitor |
[2]
|
| Hypoxanthine-guanine phosphoribosyltransferase |
Target Info |
Inhibitor |
[2]
|
| Carbonic anhydrase II |
Target Info |
Inhibitor |
[2]
|
| Delta-aminolevulinic acid dehydratase |
Target Info |
Inhibitor |
[2]
|
| Peptide deformylase |
Target Info |
Inhibitor |
[2]
|
| Heme oxygenase |
Target Info |
Inhibitor |
[2]
|
| Glutathione S-transferase |
Target Info |
Inhibitor |
[2]
|
| Heparin-binding growth factor1 |
Target Info |
Inhibitor |
[2]
|
|
BioCyc Pathway
|
Citrulline-nitric oxide cyclePWY-5920:Heme biosynthesis
|
|
Tetrapyrrole biosynthesisPWY-5874:Heme degradationPWY66-374:C20 prostanoid biosynthesis
|
|
KEGG Pathway
|
Arginine and proline metabolism
|
|
Metabolic pathways
|
|
Calcium signaling pathway
|
|
Phagosome
|
|
Circadian entrainment
|
|
Long-term depression
|
|
Salivary secretion
|
|
Alzheimer's disease
|
|
Amyotrophic lateral sclerosis (ALS)hsa00230:Purine metabolism
|
|
Drug metabolism - other enzymes
|
|
Metabolic pathwayshsa00910:Nitrogen metabolism
|
|
Proximal tubule bicarbonate reclamation
|
|
Collecting duct acid secretion
|
|
Gastric acid secretion
|
|
Pancreatic secretion
|
|
Bile secretionhsa00860:Porphyrin and chlorophyll metabolism
|
|
Metabolic pathwayshsa00860:Porphyrin and chlorophyll metabolism
|
|
HIF-1 signaling pathway
|
|
Mineral absorption
|
|
MicroRNAs in cancerhsa00590:Arachidonic acid metabolism
|
|
Metabolic pathwayshsa04010:MAPK signaling pathway
|
|
Ras signaling pathway
|
|
Rap1 signaling pathway
|
|
PI3K-Akt signaling pathway
|
|
Hippo signaling pathway
|
|
Regulation of actin cytoskeleton
|
|
Pathways in cancer
|
|
Melanoma
|
|
NetPath Pathway
|
EGFR1 Signaling PathwayNetPath_16:IL4 Signaling Pathway
|
|
EGFR1 Signaling Pathway
|
|
PANTHER Pathway
|
CCKR signaling map STP02746:Heme biosynthesisP00005:Angiogenesis
|
|
FGF signaling pathway
|
|
Pathway Interaction Database
|
Validated transcriptional targets of AP1 family members Fra1 and Fra2
|
|
HIF-1-alpha transcription factor networkfgf_pathway:FGF signaling pathway
|
|
PathWhiz Pathway
|
Arginine and Proline MetabolismPW000052:Purine MetabolismPW000158:Porphyrin MetabolismPW000158:Porphyrin Metabolism
|
|
Reactome
|
ROS production in response to bacteria
|
|
Nitric oxide stimulates guanylate cyclaseR-HSA-74217:Purine salvageR-HSA-1237044:Erythrocytes take up carbon dioxide and release oxygen
|
|
Erythrocytes take up oxygen and release carbon dioxide
|
|
Reversible hydration of carbon dioxideR-HSA-917937:Iron uptake and transportR-HSA-109704:PI3K Cascade
|
|
PIP3 activates AKT signaling
|
|
FGFR4 ligand binding and activation
|
|
FGFR3c ligand binding and activation
|
|
FGFR2c ligand binding and activation
|
|
FGFR2b ligand binding and activation
|
|
FGFR3 mutant receptor activation
|
|
Activated point mutants of FGFR2
|
|
Constitutive Signaling by Aberrant PI3K in Cancer
|
|
Phospholipase C-mediated cascade: FGFR1
|
|
Phospholipase C-mediated cascade
|
|
Phospholipase C-mediated cascade
|
|
Phospholipase C-mediated cascade
|
|
SHC-mediated cascade:FGFR1
|
|
PI-3K cascade:FGFR1
|
|
FRS-mediated FGFR1 signaling
|
|
PI-3K cascade:FGFR2
|
|
SHC-mediated cascade:FGFR2
|
|
FRS-mediated FGFR2 signaling
|
|
SHC-mediated cascade:FGFR3
|
|
FRS-mediated FGFR3 signaling
|
|
PI-3K cascade:FGFR3
|
|
FRS-mediated FGFR4 signaling
|
|
SHC-mediated cascade:FGFR4
|
|
PI-3K cascade:FGFR4
|
|
Negative regulation of FGFR1 signaling
|
|
Negative regulation of FGFR2 signaling
|
|
Negative regulation of FGFR3 signaling
|
|
Negative regulation of FGFR4 signaling
|
|
RAF/MAP kinase cascade
|
|
WikiPathways
|
Monoamine Transport
|
|
Myometrial Relaxation and Contraction Pathways
|
|
Amyotrophic lateral sclerosis (ALS)
|
|
Quercetin and Nf-kB/ AP-1 Induced Cell Apoptosis
|
|
Spinal Cord Injury
|
|
Alzheimers Disease
|
|
Effects of Nitric Oxide
|
|
Serotonin Transporter ActivityWP404:Nucleotide Metabolism
|
|
Mesodermal Commitment Pathway
|
|
Endoderm Differentiation
|
|
Metabolism of nucleotidesWP2770:Reversible Hydration of Carbon Dioxide
|
|
Uptake of Carbon Dioxide and Release of Oxygen by Erythrocytes
|
|
Uptake of Oxygen and Release of Carbon Dioxide by ErythrocytesWP561:Heme Biosynthesis
|
|
Metabolism of porphyrinsWP408:Oxidative Stress
|
|
Transcriptional activation by NRF2
|
|
NRF2 pathway
|
|
Nuclear Receptors Meta-Pathway
|
|
miR-targeted genes in squamous cell - TarBase
|
|
miR-targeted genes in muscle cell - TarBase
|
|
miR-targeted genes in lymphocytes - TarBase
|
|
miR-targeted genes in leukocytes - TarBase
|
|
miR-targeted genes in epithelium - TarBaseWP2650:Arachidonic acid metabolism
|
|
Aryl Hydrocarbon Receptor
|
|
Integrated Pancreatic Cancer PathwayWP51:Regulation of Actin Cytoskeleton
|
|
Differentiation Pathway
|
|
Hair Follicle Development: Organogenesis (Part 2 of 3)
|
|
Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R)
|
|
PIP3 activates AKT signaling
|
|
Integrated Pancreatic Cancer Pathway
|
|
Signaling by FGFR
|
| References |
| REF 1 | (http://www.guidetopharmacology.org/) Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4540). |
|---|
| REF 2 | How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. |
|---|