Drug Information
| Drug General Information | Top | |||
|---|---|---|---|---|
| Drug ID |
D0X6MT
|
|||
| Former ID |
DNC003558
|
|||
| Drug Name |
METHIOTHEPIN
|
|||
| Synonyms |
methiothepin; Metitepine; Methiothepine; Metitepinum; Metitepina; Metitepine [INN]; Metitepinum [INN-Latin]; Metitepina [INN-Spanish]; BRN 0626221; 20229-30-5; (+-)-10-(4-Methylpiperazinyl)-8-(methylthio)-10,11-dihydrodibenzo(b,f)thiepin; (+-)-8-Methylthio-10-(4-methylpiperazino)-10,11-dihydrodibenzo(b,f)thiepin; (+-)-1-(10,11-Dihydro-8-(methylthio)dibenzo(b,f)thiepin-10-yl)-4-methylpiperazine; CHEBI:64203; 1-(10,11-Dihydro-8-(methylthio)dibenzo(b,f)thiepin-10-yl)-4-methylpiperazine; NCGC00024665-03; DSSTox_CID_24000
Click to Show/Hide
|
|||
| Drug Type |
Small molecular drug
|
|||
| Indication | Discovery agent [ICD-11: N.A.] | Investigative | [1] | |
| Structure |
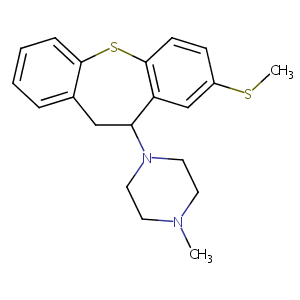 |
Download2D MOL |
||
| Formula |
C20H24N2S2
|
|||
| Canonical SMILES |
CN1CCN(CC1)C2CC3=CC=CC=C3SC4=C2C=C(C=C4)SC
|
|||
| InChI |
1S/C20H24N2S2/c1-21-9-11-22(12-10-21)18-13-15-5-3-4-6-19(15)24-20-8-7-16(23-2)14-17(18)20/h3-8,14,18H,9-13H2,1-2H3
|
|||
| InChIKey |
RLJFTICUTYVZDG-UHFFFAOYSA-N
|
|||
| CAS Number |
CAS 20229-30-5
|
|||
| PubChem Compound ID | ||||
| PubChem Substance ID |
7816804, 7979277, 8152575, 11336088, 11361327, 11372522, 11374642, 11406693, 11462299, 11466403, 11467523, 11485829, 11486186, 11489783, 11491417, 11492906, 12012717, 14828076, 24263002, 26751783, 29223214, 47216839, 47662350, 47736555, 47810816, 47810817, 47885476, 47885477, 48035204, 48110512, 48334574, 49698953, 50012693, 50104468, 50104469, 50104470, 50256032, 53790360, 57322144, 85209172, 85787800, 85788848, 90341652, 92308987, 103046125, 103307762, 104015431, 104305443, 109612645, 124750013
|
|||
| ChEBI ID |
CHEBI:64203
|
|||
| Interaction between the Drug and Microbe | Top | |||
|---|---|---|---|---|
| The Metabolism of Drug Affected by Studied Microbe(s) | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Bacteroides uniformis ATCC 8492
Show/Hide Hierarchy
|
[2] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Metitepine maleate can be metabolized by Bacteroides uniformis ATCC 8492 (log2FC = -0.765; p = 0.021). | |||
| The Abundace of Studied Microbe(s) Regulated by Drug | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Odoribacter splanchnicus
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Odoribacter splanchnicus was decreased by Methiothepin maleate (adjusted p-values: 4.23E-03). | |||
|
Studied Microbe: Parabacteroides distasonis
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Parabacteroides distasonis was decreased by Methiothepin maleate (adjusted p-values: 4.05E-03). | |||
|
Studied Microbe: Prevotella copri
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Prevotella copri was decreased by Methiothepin maleate (adjusted p-values: 2.63E-04). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bifidobacteriales | ||||
|
Studied Microbe: Bifidobacterium longum
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Bifidobacterium longum was decreased by Methiothepin maleate (adjusted p-values: 9.78E-04). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Coriobacteriales | ||||
|
Studied Microbe: Collinsella aerofaciens
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Collinsella aerofaciens was decreased by Methiothepin maleate (adjusted p-values: 3.24E-03). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Eubacteriales | ||||
|
Studied Microbe: Blautia obeum
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Blautia obeum was decreased by Methiothepin maleate (adjusted p-values: 2.50E-04). | |||
|
Studied Microbe: Eubacterium eligens
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Eubacterium eligens was decreased by Methiothepin maleate (adjusted p-values: 5.88E-04). | |||
|
Studied Microbe: Eubacterium rectale
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Eubacterium rectale was decreased by Methiothepin maleate (adjusted p-values: 1.93E-03). | |||
|
Studied Microbe: Roseburia hominis
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Roseburia hominis was decreased by Methiothepin maleate (adjusted p-values: 2.71E-04). | |||
|
Studied Microbe: Roseburia intestinalis
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Abundance Change | Decrease | |||
| Experiment Method | High-throughput screening | |||
| Description | The abundance of Roseburia intestinalis was decreased by Methiothepin maleate (adjusted p-values: 6.73E-04). | |||
| Target and Pathway | Top | |||
|---|---|---|---|---|
| Target(s) | 5-HT 5A receptor (HTR5A) | Target Info | Inhibitor | [4] |
| 5-HT 6 receptor (HTR6) | Target Info | Inhibitor | [5] | |
| 5-HT 7 receptor (HTR7) | Target Info | Inhibitor | [6] | |
| KEGG Pathway | Ras signaling pathway | |||
| Calcium signaling pathway | ||||
| Neuroactive ligand-receptor interaction | ||||
| Serotonergic synapse | ||||
| cAMP signaling pathway | ||||
| Panther Pathway | Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | |||
| Pathwhiz Pathway | Excitatory Neural Signalling Through 5-HTR 7 and Serotonin | |||
| Excitatory Neural Signalling Through 5-HTR 6 and Serotonin | ||||
| Reactome | Serotonin receptors | |||
| G alpha (s) signalling events | ||||
| G alpha (i) signalling events | ||||
| WikiPathways | Serotonin Receptor 4/6/7 and NR3C Signaling | |||
| Monoamine GPCRs | ||||
| GPCRs, Class A Rhodopsin-like | ||||
| GPCR ligand binding | ||||
| GPCR downstream signaling | ||||
| GPCRs, Other | ||||
| References | Top | |||
|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 89). | |||
| REF 2 | Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019 Jun;570(7762):462-467. | |||
| REF 3 | Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018 Mar 29;555(7698):623-628. | |||
| REF 4 | Higher-end serotonin receptors: 5-HT(5), 5-HT(6), and 5-HT(7). J Med Chem. 2003 Jul 3;46(14):2795-812. | |||
| REF 5 | 5-Cyclic amine-3-arylsulfonylindazoles as novel 5-HT6 receptor antagonists. J Med Chem. 2010 Mar 25;53(6):2521-7. | |||
| REF 6 | Novel quinazolinone derivatives as 5-HT7 receptor ligands. Bioorg Med Chem. 2008 Mar 1;16(5):2570-8. | |||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

