Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T15571
(Former ID: TTDR00565)
|
|||||
| Target Name |
5-HT 5A receptor (HTR5A)
|
|||||
| Synonyms |
Serotonin receptor 5A; 5-hydroxytryptamine receptor 5A; 5-HT5A; 5-HT-5A; 5-HT-5; 5-HT 5A
Click to Show/Hide
|
|||||
| Gene Name |
HTR5A
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Pituitary gland disorder [ICD-11: 5A60-5A61] | |||||
| Function |
The activity of this receptor is mediated by G proteins. This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen.
Click to Show/Hide
|
|||||
| BioChemical Class |
GPCR rhodopsin
|
|||||
| UniProt ID | ||||||
| Sequence |
MDLPVNLTSFSLSTPSPLETNHSLGKDDLRPSSPLLSVFGVLILTLLGFLVAATFAWNLL
VLATILRVRTFHRVPHNLVASMAVSDVLVAALVMPLSLVHELSGRRWQLGRRLCQLWIAC DVLCCTASIWNVTAIALDRYWSITRHMEYTLRTRKCVSNVMIALTWALSAVISLAPLLFG WGETYSEGSEECQVSREPSYAVFSTVGAFYLPLCVVLFVYWKIYKAAKFRVGSRKTNSVS PISEAVEVKDSAKQPQMVFTVRHATVTFQPEGDTWREQKEQRAALMVGILIGVFVLCWIP FFLTELISPLCSCDIPAIWKSIFLWLGYSNSFFNPLIYTAFNKNYNSAFKNFFSRQH Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Metergolin | Drug Info | Approved | Hyperprolactinaemia | [2] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Antagonist | [+] 5 Antagonist drugs | + | ||||
| 1 | Metergolin | Drug Info | [1] | |||
| 2 | bufotenine | Drug Info | [6] | |||
| 3 | MPDT | Drug Info | [7] | |||
| 4 | SB 699551 | Drug Info | [9] | |||
| 5 | SB-699551-A | Drug Info | [10] | |||
| Inhibitor | [+] 16 Inhibitor drugs | + | ||||
| 1 | (+/-)-nantenine | Drug Info | [3] | |||
| 2 | 3,4-dihydroquinazolin-2-amine hydrobromide | Drug Info | [4] | |||
| 3 | 4-ethyl-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 4 | 4-methyl-N-propyl-3,4-dihydroquinazolin-2-amine | Drug Info | [5] | |||
| 5 | 4-propyl-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 6 | 5,6-dichloro-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 7 | 5-chloro-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 8 | 5-chloro-4-ethyl-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 9 | 5-chloro-4-methyl-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 10 | 8-chloro-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 11 | 8-methoxy-4-methyl-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 12 | METHIOTHEPIN | Drug Info | [8] | |||
| 13 | N,4-dimethyl-3,4-dihydroquinazolin-2-amine | Drug Info | [5] | |||
| 14 | N,N-dimethyl-3,4-dihydroquinazolin-2-amine | Drug Info | [4] | |||
| 15 | N-butyl-4-methyl-3,4-dihydroquinazolin-2-amine | Drug Info | [5] | |||
| 16 | SEROTONIN | Drug Info | [11] | |||
| Agonist | [+] 6 Agonist drugs | + | ||||
| 1 | 5-CT | Drug Info | [6] | |||
| 2 | EDMT | Drug Info | [7] | |||
| 3 | lysergic acid | Drug Info | [6] | |||
| 4 | TFMPP | Drug Info | [12] | |||
| 5 | [125I]LSD | Drug Info | [13] | |||
| 6 | [3H]5-CT | Drug Info | [13] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Methylergonovine | Ligand Info | |||||
| Structure Description | CryoEM structure of Go-coupled 5-HT5AR in complex with Methylergometrine | PDB:7UM7 | ||||
| Method | Electron microscopy | Resolution | 2.75 Å | Mutation | No | [14] |
| PDB Sequence |
GVLILTLLGF
49 LVAATFAWNL59 LVLATILRVR69 TFHRVPHNLV79 ASMAVSDVLV89 AALVMPLSLV 99 HELSGRRWQL109 GRRLCQLWIA119 CDVLCCTASI129 WNVTAIALDR139 YWSITRPMEY 149 TLRTRKCVSN159 VMIALTWALS169 AVISLAPLLF179 GEECQVSREP198 SYAVFSTVGA 208 FYLPLCVVLF218 VYWKIYKAAK228 FRVEQKEQRA283 ALMVGILIGV293 FVLCWIPFFL 303 TELISPLCSC313 DIPAIWKSIF323 LWLGYSNSFF333 NPLIYTAFNK343 NYNSAF |
|||||
|
|
||||||
| Ligand Name: Lisuride | Ligand Info | |||||
| Structure Description | CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride | PDB:7UM6 | ||||
| Method | Electron microscopy | Resolution | 2.79 Å | Mutation | Yes | [14] |
| PDB Sequence |
GVLILTLLGF
49 LVAATFAWNL59 LVLATILRVR69 TFHRVPHNLV79 ASMAVSDVLV89 AALVMPLSLV 99 HELSGRRWQL109 GRRLCQLWIA119 CDVLCCTASI129 WNVTAIALDR139 YWSITRPMEY 149 TLRTRKCVSN159 VMIALTWALS169 AVISLAPLLF179 GEECQVSREP198 SYAVFSTVGA 208 FYLPLCVVLF218 VYWKIYKAAK228 FRVWREQKEQ281 RAALMVGILI291 GVFVLCWIPF 301 FLTELISPLC311 SCDIPAIWKS321 IFLWLGYSNS331 FFNPLIYTAF341 NKNYNSAF |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Pathway Affiliation
|
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
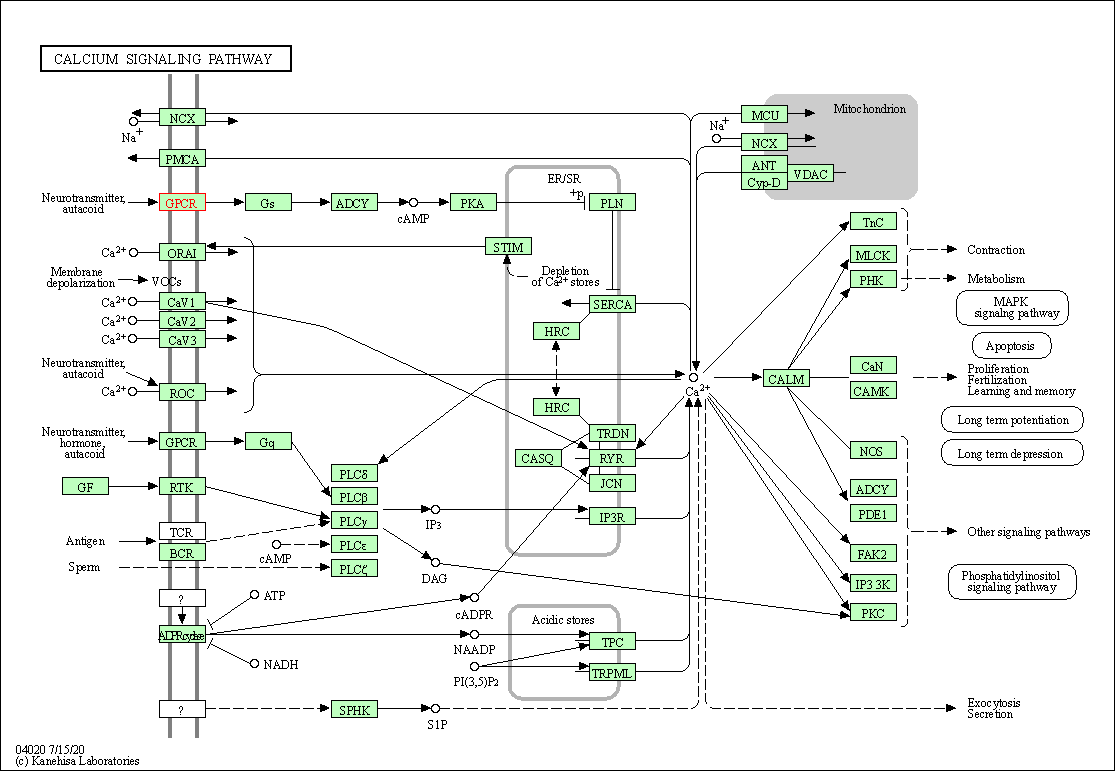
| Calcium signaling pathway | hsa04020 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
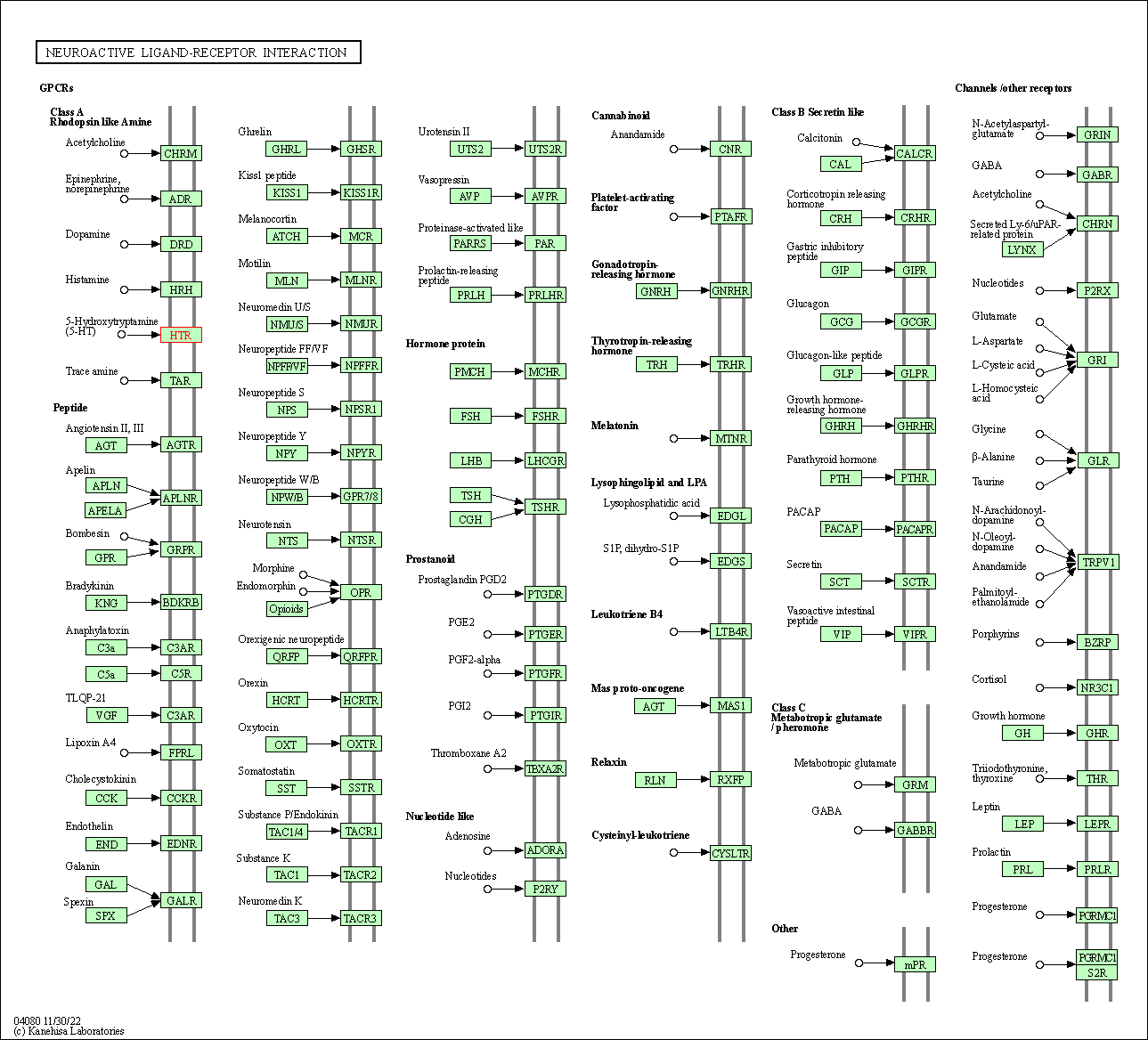
| Neuroactive ligand-receptor interaction | hsa04080 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
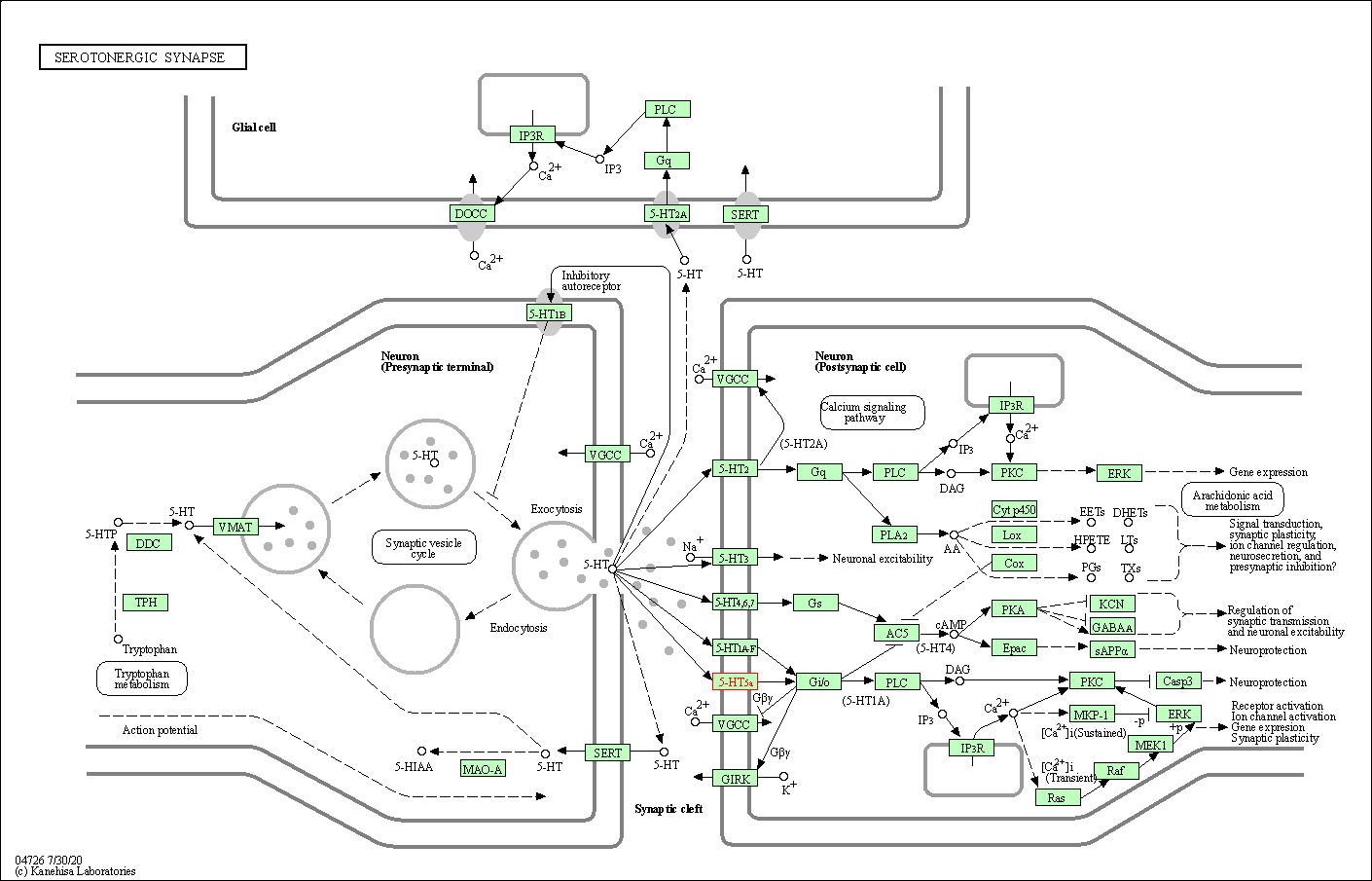
| Serotonergic synapse | hsa04726 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Calcium signaling pathway | |||||
| 2 | Neuroactive ligand-receptor interaction | |||||
| 3 | Serotonergic synapse | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Serotonin receptors | |||||
| 2 | G alpha (i) signalling events | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Monoamine GPCRs | |||||
| 2 | GPCRs, Class A Rhodopsin-like | |||||
| 3 | GPCR ligand binding | |||||
| 4 | GPCR downstream signaling | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Cloning and characterisation of the human 5-HT5A serotonin receptor. FEBS Lett. 1994 Dec 5;355(3):242-6. | |||||
| REF 2 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 3 | Synthetic studies and pharmacological evaluations on the MDMA ('Ecstasy') antagonist nantenine. Bioorg Med Chem Lett. 2010 Jan 15;20(2):628-31. | |||||
| REF 4 | Cyclic guanidines as dual 5-HT5A/5-HT7 receptor ligands: structure-activity relationship elucidation. Bioorg Med Chem Lett. 2008 Jan 1;18(1):256-61. | |||||
| REF 5 | Cyclic guanidines as dual 5-HT5A/5-HT7 receptor ligands: optimising brain penetration. Bioorg Med Chem Lett. 2008 Jan 1;18(1):262-6. | |||||
| REF 6 | Expression of functional mouse 5-HT5A serotonin receptor in the methylotrophic yeast Pichia pastoris: pharmacological characterization and localization. FEBS Lett. 1995 Dec 27;377(3):451-6. | |||||
| REF 7 | 2-Substituted tryptamines: agents with selectivity for 5-HT(6) serotonin receptors. J Med Chem. 2000 Mar 9;43(5):1011-8. | |||||
| REF 8 | Higher-end serotonin receptors: 5-HT(5), 5-HT(6), and 5-HT(7). J Med Chem. 2003 Jul 3;46(14):2795-812. | |||||
| REF 9 | Discovery of a potent and selective 5-ht5A receptor antagonist by high-throughput chemistry. Bioorg Med Chem Lett. 2005 Sep 15;15(18):4014-8. | |||||
| REF 10 | 5-ht5A receptors as a therapeutic target. Pharmacol Ther. 2006 Sep;111(3):707-14. | |||||
| REF 11 | Identification of a potent, selective, and orally active leukotriene a4 hydrolase inhibitor with anti-inflammatory activity. J Med Chem. 2008 Jul 24;51(14):4150-69. | |||||
| REF 12 | Mouse 5-hydroxytryptamine5A and 5-hydroxytryptamine5B receptors define a new family of serotonin receptors: cloning, functional expression, and chromosomal localization. Mol Pharmacol. 1993 Mar;43(3):313-9. | |||||
| REF 13 | Human 5-HT(5) receptors: the 5-HT(5A) receptor is functional but the 5-HT(5B) receptor was lost during mammalian evolution. Eur J Pharmacol. 2001 Apr 27;418(3):157-67. | |||||
| REF 14 | Inactive and active state structures template selective tools for the human 5-HT(5A) receptor. Nat Struct Mol Biol. 2022 Jul;29(7):677-687. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

