Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T01851
(Former ID: TTDC00148)
|
|||||
| Target Name |
Integrin beta-1 (ITGB1)
|
|||||
| Synonyms |
VLA-4 subunit beta; MSK12; MDF2; Integrin VLA-4 beta subunit; Glycoprotein IIa; GPIIA; Fibronectin receptor subunit beta; Fibronectin receptor beta subunit; FNRB; CD29 antigen; CD29; Beta(1) integrin
Click to Show/Hide
|
|||||
| Gene Name |
ITGB1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Lung cancer [ICD-11: 2C25] | |||||
| 2 | Brain cancer [ICD-11: 2A00] | |||||
| 3 | Retinopathy [ICD-11: 9B71] | |||||
| Function |
Integrins alpha-1/beta-1 and alpha-2/beta-2 recognize the proline-hydroxylated sequence G-F-P-G-E-R in collagen. Integrins alpha-2/beta-1, alpha-3/beta-1, alpha-4/beta-1, alpha-5/beta-1, alpha-8/beta-1, alpha-10/beta-1, alpha-11/beta-1 and alpha-V/beta-1 are receptors for fibronectin. Alpha-4/beta-1 recognizes one or more domains within the alternatively spliced CS-1 and CS-5 regions of fibronectin. Integrin alpha-5/beta-1 is a receptor for fibrinogen. Integrin alpha-1/beta-1, alpha-2/beta-1, alpha-6/beta-1 and alpha-7/beta-1 are receptors for lamimin. Integrin alpha-6/beta-1 (ITGA6:ITGB1) is present in oocytes and is involved in sperm-egg fusion. Integrin alpha-4/beta-1 is a receptor for VCAM1. It recognizes the sequence Q-I-D-S in VCAM1. Integrin alpha-9/beta-1 is a receptor for VCAM1, cytotactin and osteopontin. It recognizes the sequence A-E-I-D-G-I-E-L in cytotactin. Integrin alpha-3/beta-1 is a receptor for epiligrin, thrombospondin and CSPG4. Alpha-3/beta-1 may mediate with LGALS3 the stimulation by CSPG4 of endothelial cells migration. Integrin alpha-V/beta-1 is a receptor for vitronectin. Beta-1 integrins recognize the sequence R-G-D in a wide array of ligands. Isoform 2 interferes with isoform 1 resulting in a dominant negative effect on cell adhesion and migration (in vitro). When associated with alpha-7/beta-1 integrin, regulates cell adhesion and laminin matrix deposition. Involved in promoting endothelial cell motility and angiogenesis. Involved in osteoblast compaction through the fibronectin fibrillogenesis cell-mediated matrix assembly process and the formation of mineralized bone nodules. May be involved in up-regulation of the activity of kinases such as PKC via binding to KRT1. Together with KRT1 and RACK1, serves as a platform for SRC activation or inactivation. Plays a mechanistic adhesive role during telophase, required for the successful completion of cytokinesis. Integrin alpha-3/beta-1 provides a docking site for FAP (seprase) at invadopodia plasma membranes in a collagen-dependent manner and hence may participate in the adhesion, formation of invadopodia and matrix degradation processes, promoting cell invasion. ITGA4:ITGB1 binds to fractalkine (CX3CL1) and may act as its coreceptor in CX3CR1-dependent fractalkine signaling. ITGA4:ITGB1 and ITGA5:ITGB1 bind to PLA2G2A via a site (site 2) which is distinct from the classical ligand-binding site (site 1) and this induces integrin conformational changes and enhanced ligand binding to site 1. ITGA5:ITGB1 acts as a receptor for fibrillin-1 (FBN1) and mediates R-G-D-dependent cell adhesion to FBN1. ITGA5:ITGB1 is a receptor for IL1B and binding is essential for IL1B signaling. Integrins alpha-1/beta-1, alpha-2/beta-1, alpha-10/beta-1 and alpha-11/beta-1 are receptors for collagen.
Click to Show/Hide
|
|||||
| BioChemical Class |
Integrin
|
|||||
| UniProt ID | ||||||
| Sequence |
MNLQPIFWIGLISSVCCVFAQTDENRCLKANAKSCGECIQAGPNCGWCTNSTFLQEGMPT
SARCDDLEALKKKGCPPDDIENPRGSKDIKKNKNVTNRSKGTAEKLKPEDITQIQPQQLV LRLRSGEPQTFTLKFKRAEDYPIDLYYLMDLSYSMKDDLENVKSLGTDLMNEMRRITSDF RIGFGSFVEKTVMPYISTTPAKLRNPCTSEQNCTSPFSYKNVLSLTNKGEVFNELVGKQR ISGNLDSPEGGFDAIMQVAVCGSLIGWRNVTRLLVFSTDAGFHFAGDGKLGGIVLPNDGQ CHLENNMYTMSHYYDYPSIAHLVQKLSENNIQTIFAVTEEFQPVYKELKNLIPKSAVGTL SANSSNVIQLIIDAYNSLSSEVILENGKLSEGVTISYKSYCKNGVNGTGENGRKCSNISI GDEVQFEISITSNKCPKKDSDSFKIRPLGFTEEVEVILQYICECECQSEGIPESPKCHEG NGTFECGACRCNEGRVGRHCECSTDEVNSEDMDAYCRKENSSEICSNNGECVCGQCVCRK RDNTNEIYSGKFCECDNFNCDRSNGLICGGNGVCKCRVCECNPNYTGSACDCSLDTSTCE ASNGQICNGRGICECGVCKCTDPKFQGQTCEMCQTCLGVCAEHKECVQCRAFNKGEKKDT CTQECSYFNITKVESRDKLPQPVQPDPVSHCKEKDVDDCWFYFTYSVNGNNEVMVHVVEN PECPTGPDIIPIVAGVVAGIVLIGLALLLIWKLLMIIHDRREFAKFEKEKMNAKWDTGEN PIYKSAVTTVVNPKYEGK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T15VAS | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | 131I-radretumab | Drug Info | Phase 1/2 | Non-small-cell lung cancer | [2] | |
| 2 | JSM 6427 | Drug Info | Phase 1 | Macular degeneration | [3] | |
| 3 | OS2966 | Drug Info | Phase 1 | Glioblastoma of brain | [4] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Antagonist | [+] 1 Antagonist drugs | + | ||||
| 1 | JSM 6427 | Drug Info | [5], [6], [7] | |||
| Inhibitor | [+] 4 Inhibitor drugs | + | ||||
| 1 | OS2966 | Drug Info | [8] | |||
| 2 | C(-GRGDfL-) | Drug Info | [9] | |||
| 3 | MK-0668 | Drug Info | [10] | |||
| 4 | SB-265123 | Drug Info | [11] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
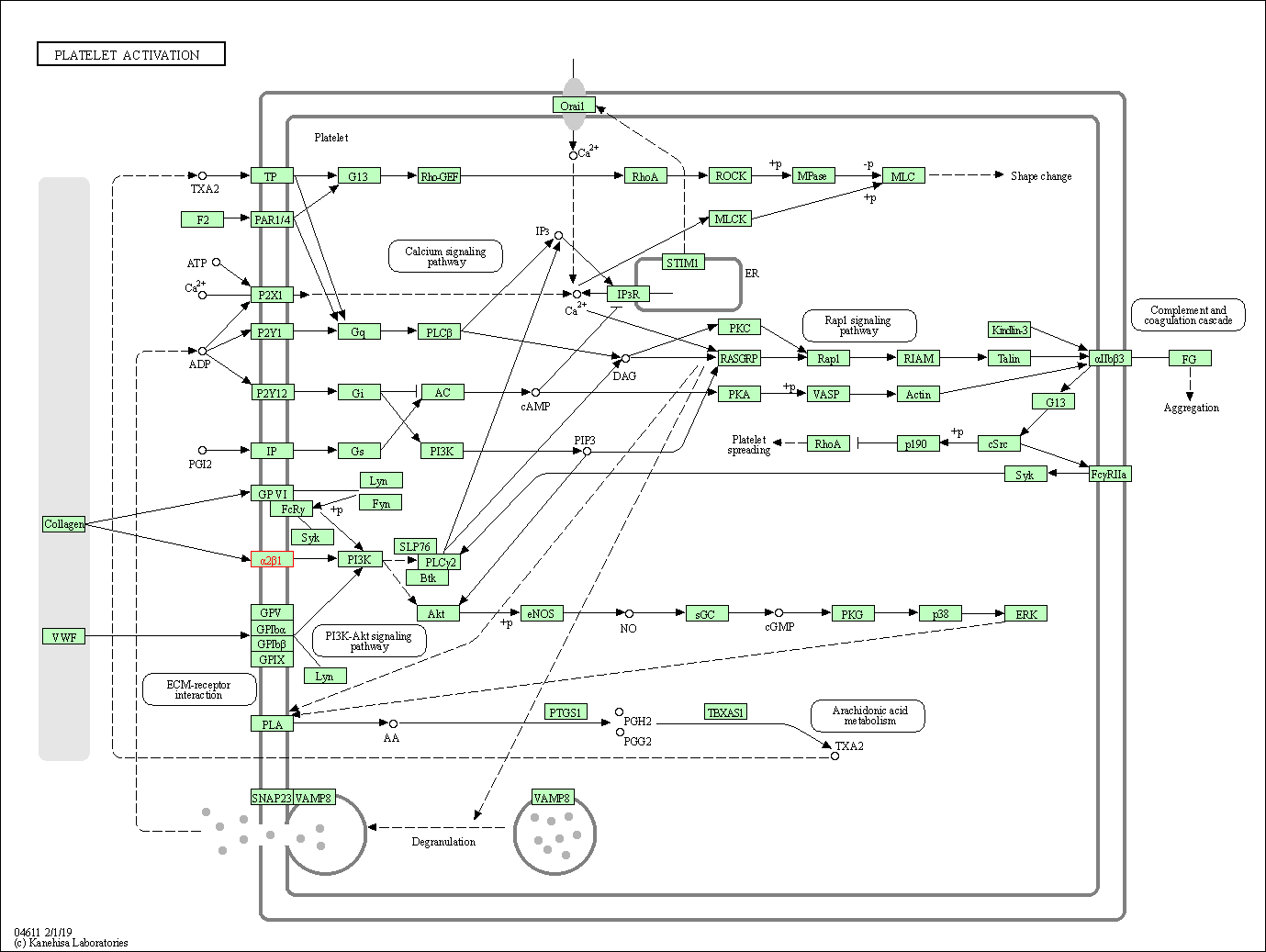
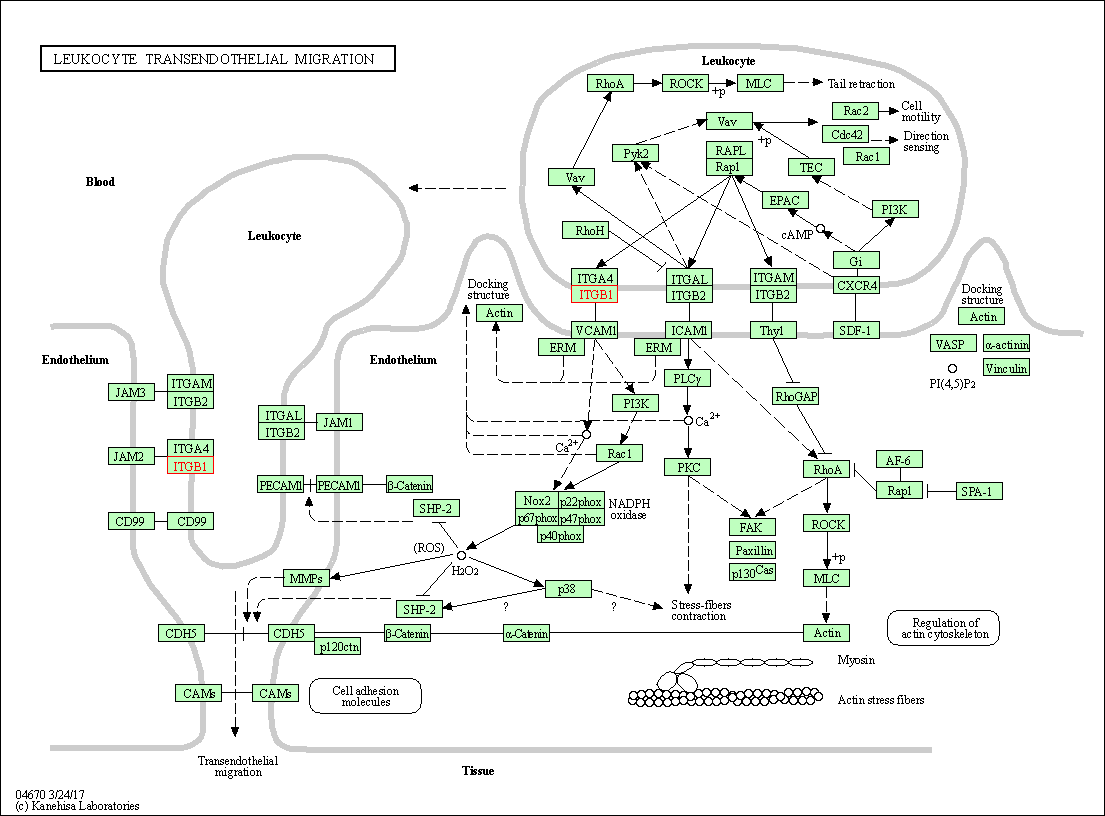
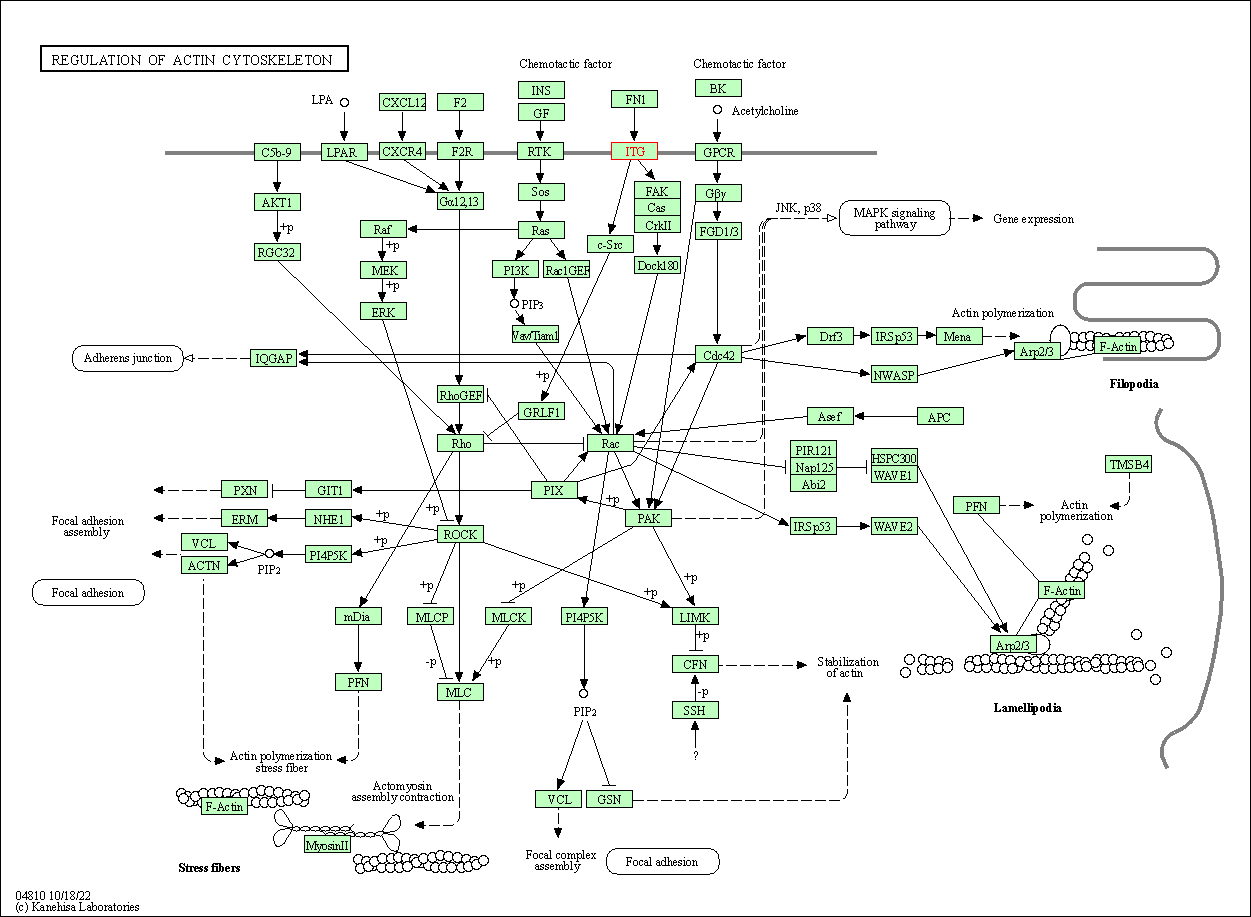
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phagosome | hsa04145 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Axon guidance | hsa04360 | Affiliated Target |

|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
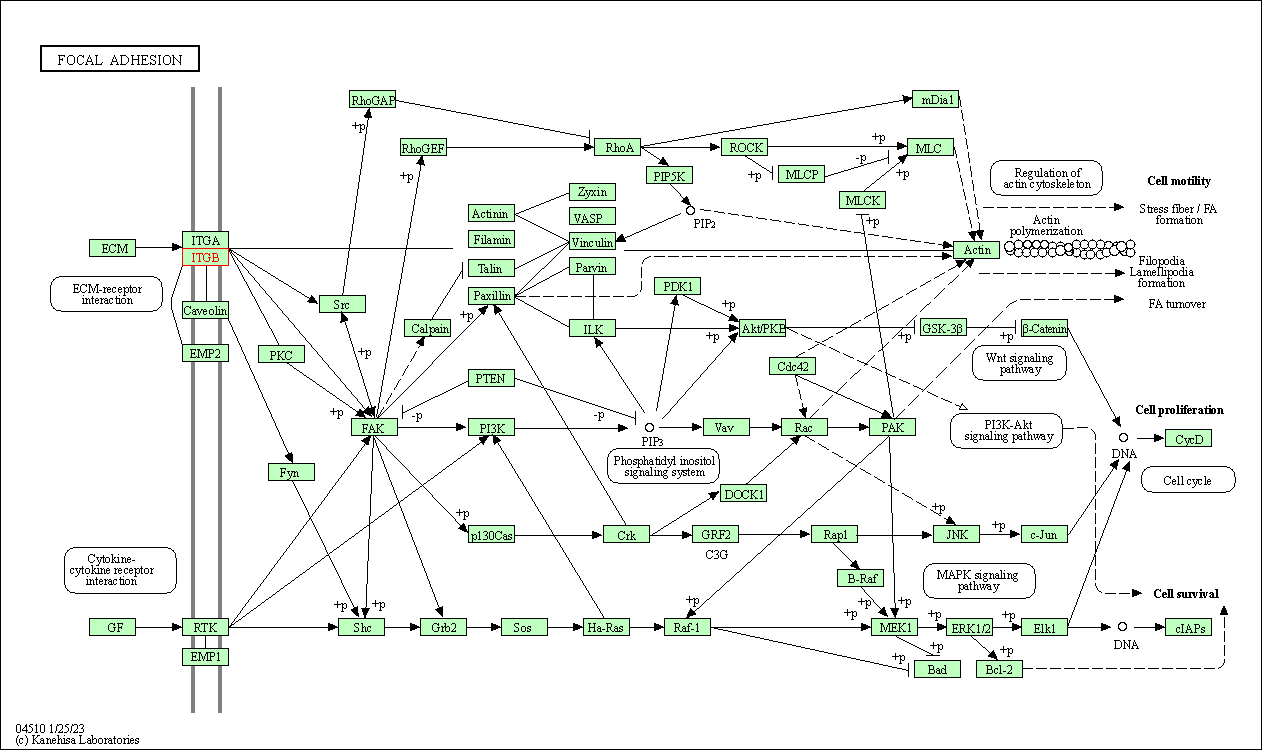
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
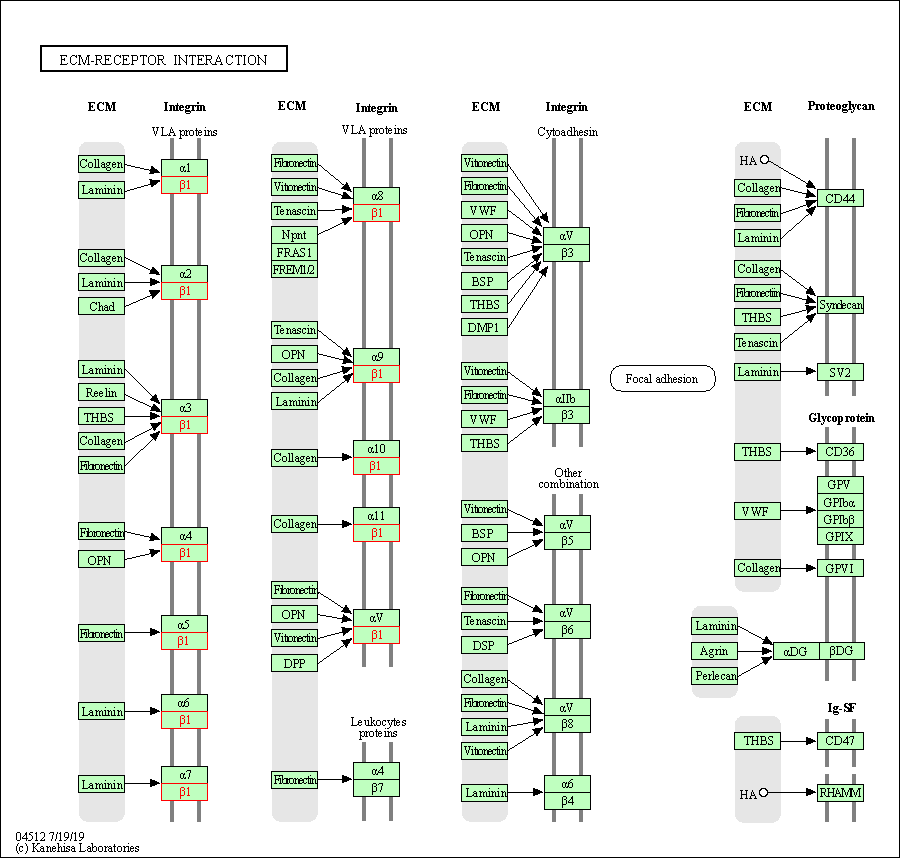
| ECM-receptor interaction | hsa04512 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
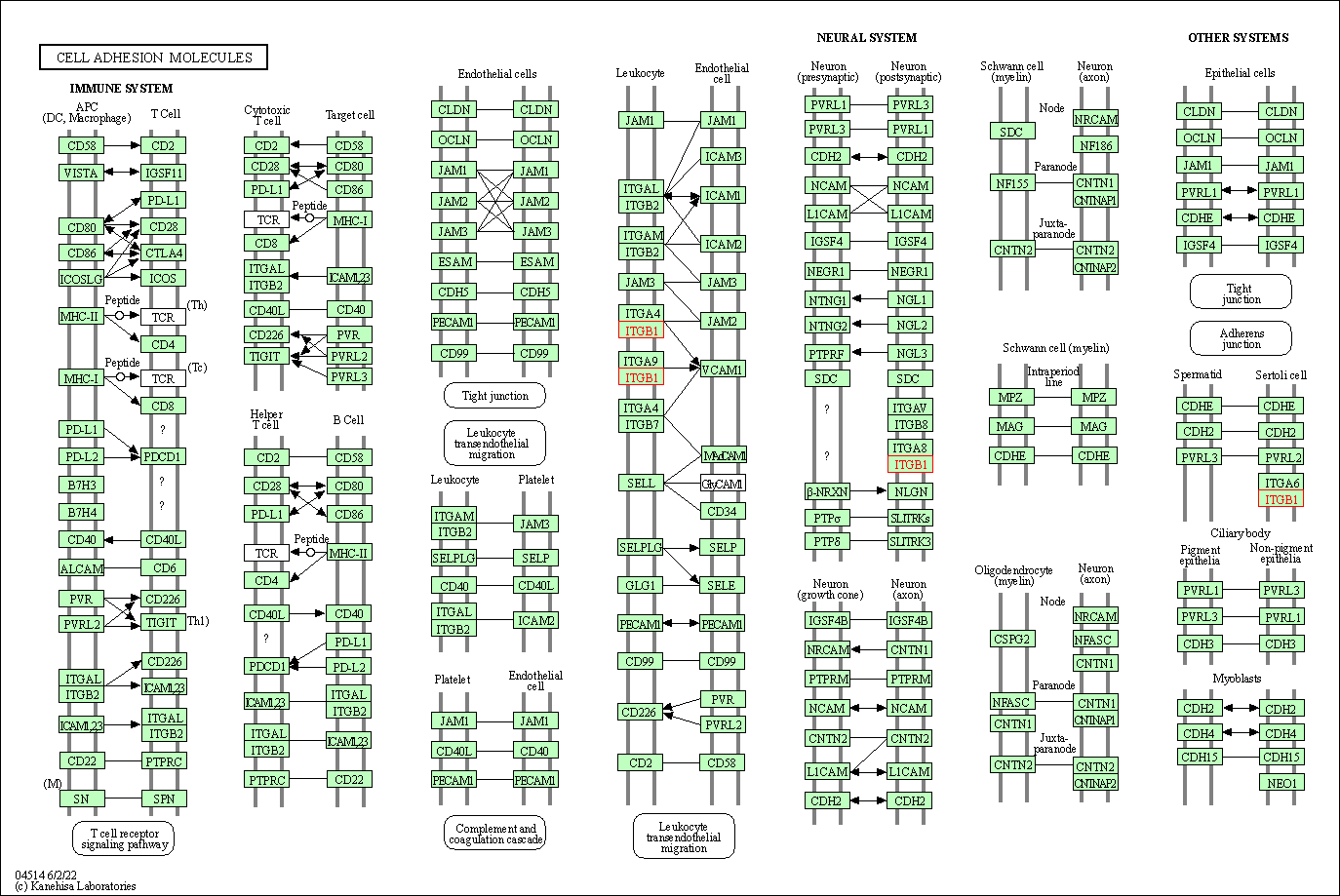
| Cell adhesion molecules | hsa04514 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
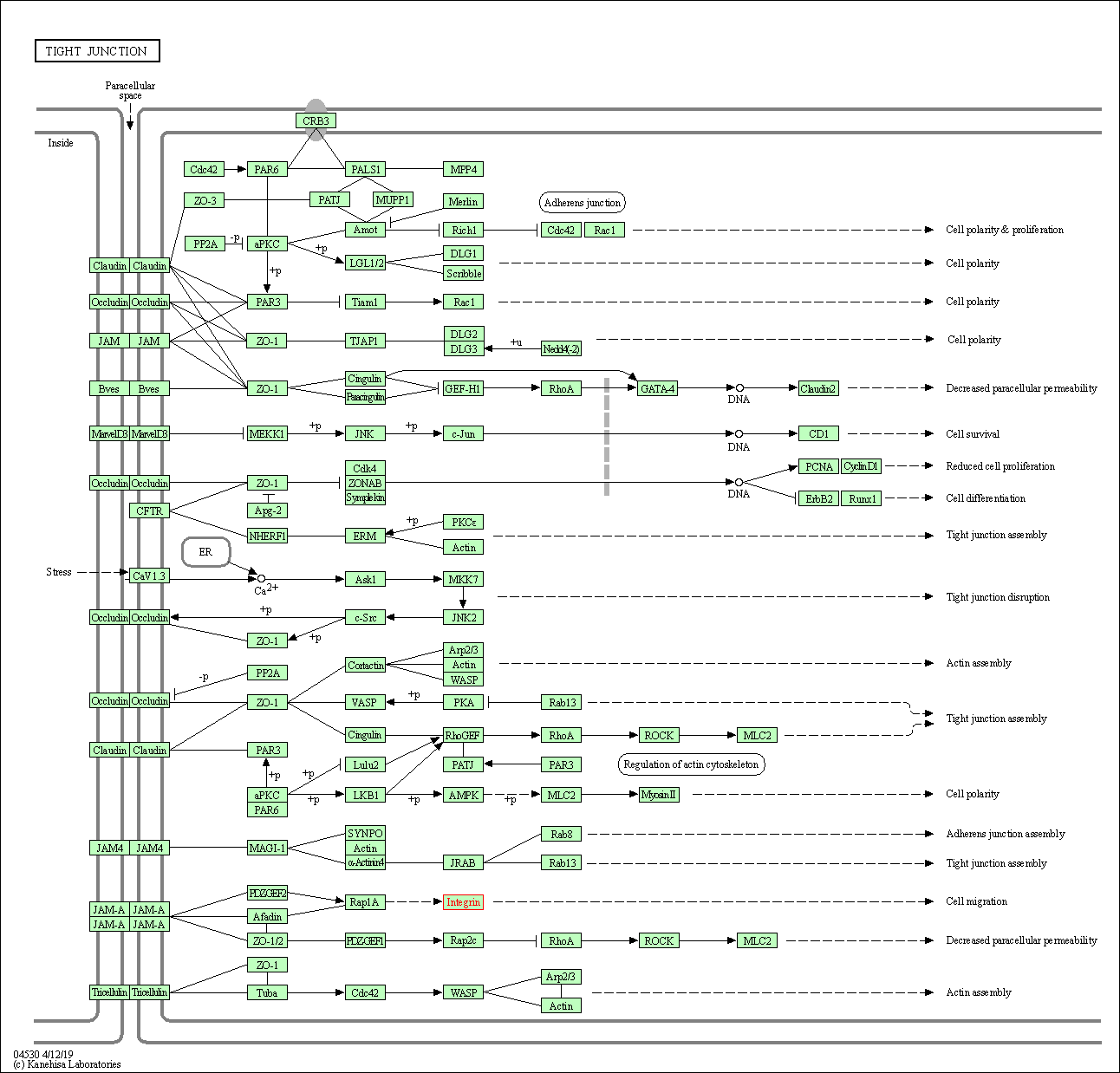
| Tight junction | hsa04530 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Leukocyte transendothelial migration | hsa04670 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 71 | Degree centrality | 7.63E-03 | Betweenness centrality | 7.34E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.50E-01 | Radiality | 1.44E+01 | Clustering coefficient | 9.05E-02 |
| Neighborhood connectivity | 2.79E+01 | Topological coefficient | 3.46E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Radretumab radioimmunotherapy in patients with brain metastasis: a 124I-L19SIP dosimetric PET study. Cancer Immunol Res. 2013 Aug;1(2):134-43. | |||||
| REF 2 | ClinicalTrials.gov (NCT01242943) Radioimmunotherapy With 131I-L19SIP in Patients With Cancer. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT00536016) A Phase 1 Safety Study of Single and Repeated Doses of JSM6427 (Intravitreal Injection) to Treat AMD. U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT04608812) Convection-enhanced Delivery of OS2966 for Patients With High-grade Glioma Undergoing a Surgical Resection. U.S. National Institutes of Health. | |||||
| REF 5 | An alpha5beta1 integrin inhibitor attenuates glioma growth. Mol Cell Neurosci. 2008 Dec;39(4):579-85. | |||||
| REF 6 | Modulation of hypoxia-induced neovascularization by JSM6427, an integrin alpha5beta1 inhibiting molecule. Curr Eye Res. 2007 Sep;32(9):801-12. | |||||
| REF 7 | Suppression and regression of choroidal neovascularization by systemic administration of an alpha5beta1 integrin antagonist. Mol Pharmacol. 2006 Jun;69(6):1820-8. | |||||
| REF 8 | Convection-Enhanced Delivery of a First-in-Class Anti-beta1 Integrin Antibody for the Treatment of High-Grade Glioma Utilizing Real-Time Imaging. Pharmaceutics. 2020 Dec 30;13(1):40. | |||||
| REF 9 | Multiple N-methylation by a designed approach enhances receptor selectivity. J Med Chem. 2007 Nov 29;50(24):5878-81. | |||||
| REF 10 | Discovery of N-{N-[(3-cyanobenzene) sulfonyl]-4(R)-(3,3-difluoropiperidin-1-yl)-(l)-prolyl}-4-[(3',5'-dichloro-isonicotinoyl) amino]-(l)-phenylalan... Bioorg Med Chem Lett. 2009 Oct 1;19(19):5803-6. | |||||
| REF 11 | Orally bioavailable nonpeptide vitronectin receptor antagonists with efficacy in an osteoporosis model. Bioorg Med Chem Lett. 1999 Jul 5;9(13):1807-12. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

