Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T13616
(Former ID: TTDR00485)
|
|||||
| Target Name |
Integrin-linked protein kinase 1 (ILK)
|
|||||
| Synonyms |
P59ILK; Integrin-linked protein kinase; Integrin-linked kinase; ILK2; ILK1; ILK-2; ILK-1; Beta-integrin-linked kinase; 59 kDa serine/threonine-protein kinase; 59 kDa serine/threonine protein kinase
Click to Show/Hide
|
|||||
| Gene Name |
ILK
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
May act as a mediator of inside-out integrin signaling. Focal adhesion protein part of the complex ILK-PINCH. This complex is considered to be one of the convergence points of integrin- and growth factor-signaling pathway. Could be implicated in mediating cell architecture, adhesion to integrin substrates and anchorage-dependent growth in epithelial cells. Phosphorylates beta-1 and beta-3 integrin subunit on serine and threonine residues, but also AKT1 and GSK3B. Receptor-proximal protein kinase regulating integrin-mediated signal transduction.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MDDIFTQCREGNAVAVRLWLDNTENDLNQGDDHGFSPLHWACREGRSAVVEMLIMRGARI
NVMNRGDDTPLHLAASHGHRDIVQKLLQYKADINAVNEHGNVPLHYACFWGQDQVAEDLV ANGALVSICNKYGEMPVDKAKAPLRELLRERAEKMGQNLNRIPYKDTFWKGTTRTRPRNG TLNKHSGIDFKQLNFLTKLNENHSGELWKGRWQGNDIVVKVLKVRDWSTRKSRDFNEECP RLRIFSHPNVLPVLGACQSPPAPHPTLITHWMPYGSLYNVLHEGTNFVVDQSQAVKFALD MARGMAFLHTLEPLIPRHALNSRSVMIDEDMTARISMADVKFSFQCPGRMYAPAWVAPEA LQKKPEDTNRRSADMWSFAVLLWELVTREVPFADLSNMEIGMKVALEGLRPTIPPGISPH VCKLMKICMNEDPAKRPKFDMIVPILEKMQDK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T96YED | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Crystal structure of the ILK/alpha-parvin core complex (MnATP) | PDB:3REP | ||||
| Method | X-ray diffraction | Resolution | 1.80 Å | Mutation | Yes | [4] |
| PDB Sequence |
HSGIDFKQLN
194 FLTKLNENHS204 GELWKGRWQG214 NDIVVKVLKV224 RDWSTRKSRD234 FNEECPRLRI 244 FSHPNVLPVL254 GACQSPPAPH264 PTLITHWMPY274 GSLYNVLHEG284 TNFVVDQSQA 294 VKFALDMARG304 MAFLHTLEPL314 IPRHALNSRS324 VMIDEDMTAR334 ISMADVKFSF 344 QSPGRMYAPA354 WVAPEALQKK364 PEDTNRRSAD374 MWSFAVLLWE384 LVTREVPFAD 394 LSNMEIGMKV404 ALEGLRPTIP414 PGISPHVSKL424 MKICMNEDPA434 KRPKFDMIVP 444 ILEKMQDK
|
|||||
|
|
LEU199
4.554
ASN200
2.920
ASN202
2.985
HIS203
3.577
SER204
2.553
LEU207
2.993
VAL218
3.647
LYS220
2.683
LEU251
4.148
THR269
3.654
HIS270
2.894
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|

| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| PPAR signaling pathway | hsa03320 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
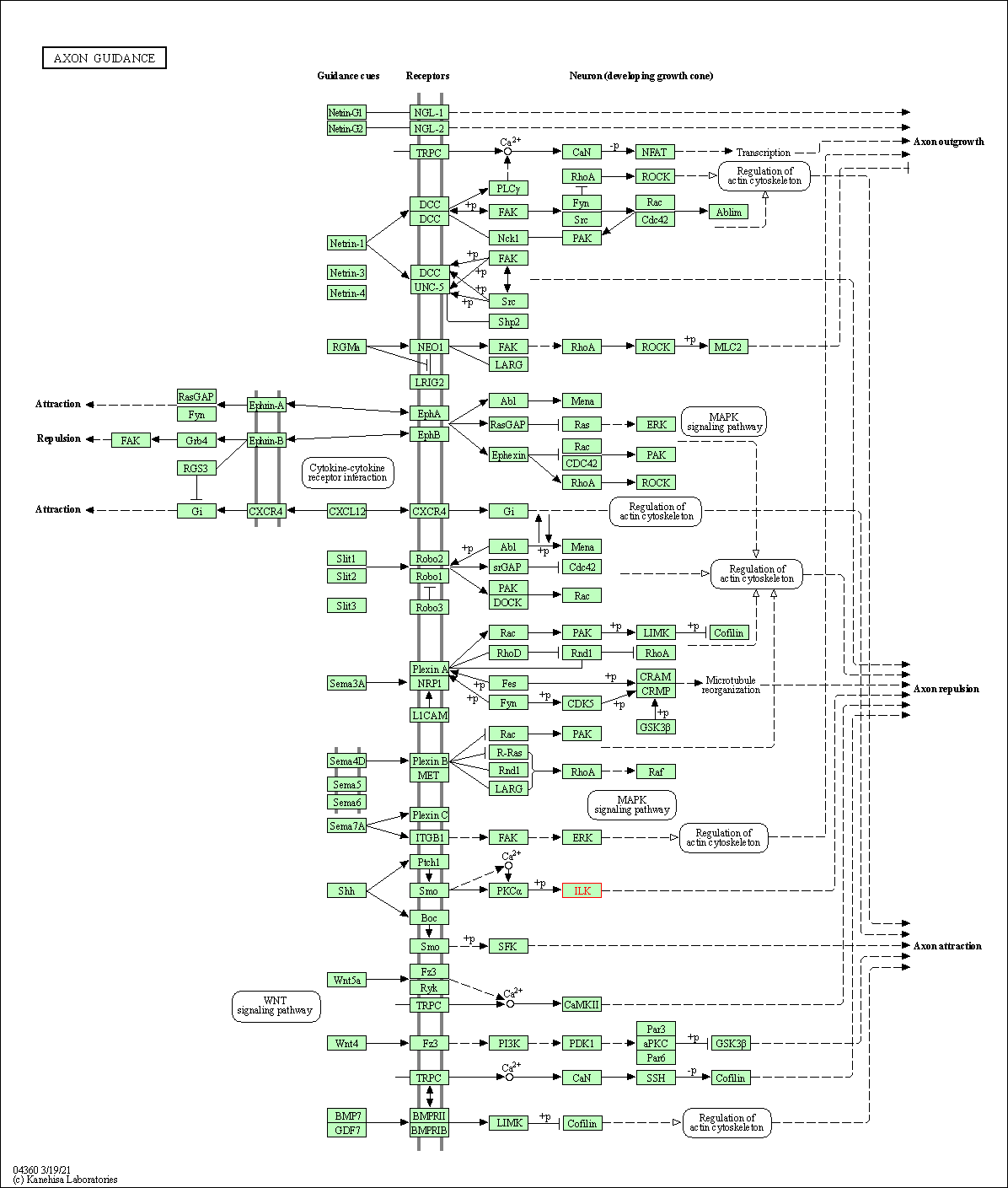
| Axon guidance | hsa04360 | Affiliated Target |

|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
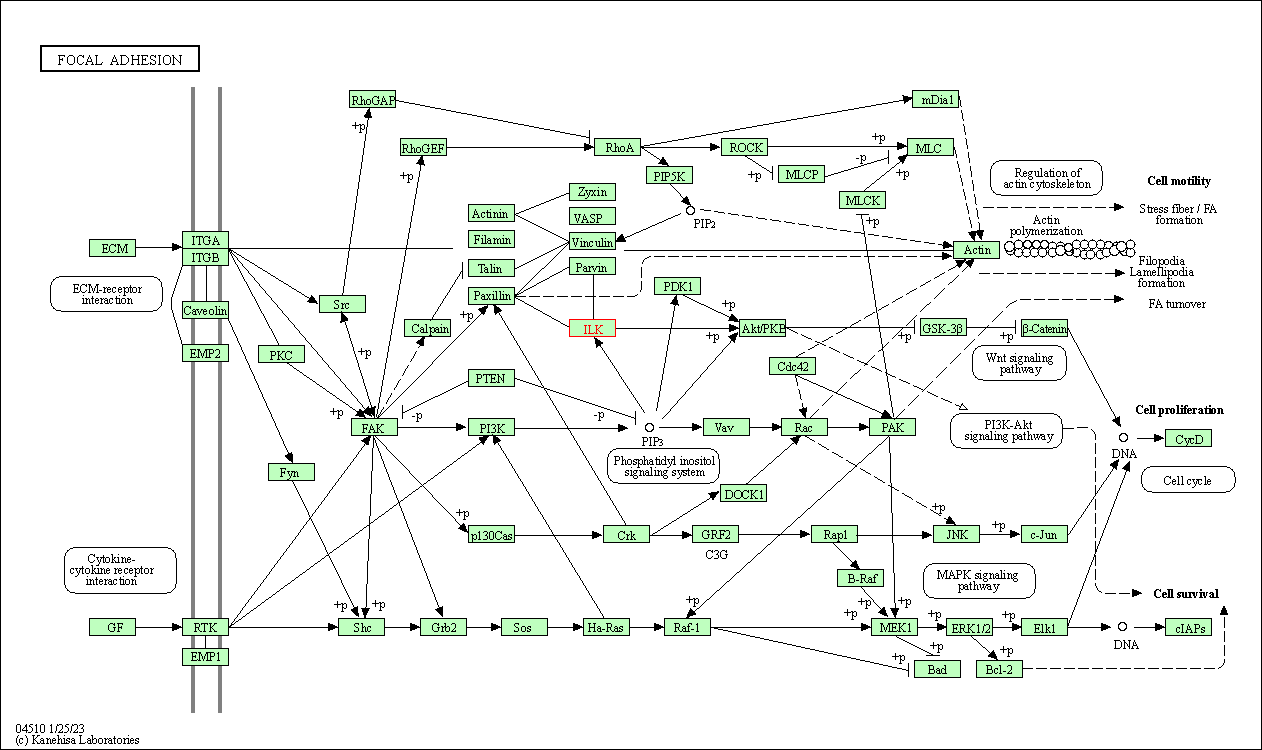
| Focal adhesion | hsa04510 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Degree | 29 | Degree centrality | 3.12E-03 | Betweenness centrality | 2.05E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.53E-01 | Radiality | 1.44E+01 | Clustering coefficient | 1.16E-01 |
| Neighborhood connectivity | 3.51E+01 | Topological coefficient | 5.50E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| NetPath Pathway | [+] 2 NetPath Pathways | + | ||||
| 1 | TCR Signaling Pathway | |||||
| 2 | FSH Signaling Pathway | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Integrin signalling pathway | |||||
| PID Pathway | [+] 3 PID Pathways | + | ||||
| 1 | Osteopontin-mediated events | |||||
| 2 | Integrin-linked kinase signaling | |||||
| 3 | Integrins in angiogenesis | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Localization of the PINCH-ILK-PARVIN complex to focal adhesions | |||||
| WikiPathways | [+] 5 WikiPathways | + | ||||
| 1 | Eukaryotic Transcription Initiation | |||||
| 2 | Focal Adhesion | |||||
| 3 | Primary Focal Segmental Glomerulosclerosis FSGS | |||||
| 4 | Integrin-mediated Cell Adhesion | |||||
| 5 | Cell junction organization | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2041). | |||||
| REF 2 | Pyrazolylbenzothiazole derivatives and their use as therapeutic agents. US8754233. | |||||
| REF 3 | Identification and characterization of a novel integrin-linked kinase inhibitor. J Med Chem. 2011 Sep 22;54(18):6364-74. | |||||
| REF 4 | Crystal structure of the ILK/alpha-parvin core complex (MnATP) | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

