Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T31721
(Former ID: TTDI03191)
|
|||||
| Target Name |
Excitatory amino acid transporter 3 (SLC1A1)
|
|||||
| Synonyms |
Solute carrier family 1 member 1; Sodium-dependent glutamate/aspartate transporter 3; Neuronal and epithelial glutamate transporter; Excitatory amino-acid carrier 1; EAAT3; EAAC1
Click to Show/Hide
|
|||||
| Gene Name |
SLC1A1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Can also transport L-cysteine. Functions as a symporter that transports one amino acid molecule together with two or three Na(+) ions and one proton, in parallel with the counter-transport of one K(+) ion. Mediates Cl(-) flux that is not coupled to amino acid transport; this avoids the accumulation of negative charges due to aspartate and Na(+) symport. Plays an important role in L-glutamate and L-aspartate reabsorption in renal tubuli. Plays a redundant role in the rapid removal of released glutamate from the synaptic cleft, which is essential for terminating the postsynaptic action of glutamate. Negatively regulated by ARL6IP5. Sodium-dependent, high-affinity amino acid transporter that mediates the uptake of L-glutamate and also L-aspartate and D-aspartate.
Click to Show/Hide
|
|||||
| BioChemical Class |
Dicarboxylate/amino acid:cation symporter
|
|||||
| UniProt ID | ||||||
| Sequence |
MGKPARKGCEWKRFLKNNWVLLSTVAAVVLGITTGVLVREHSNLSTLEKFYFAFPGEILM
RMLKLIILPLIISSMITGVAALDSNVSGKIGLRAVVYYFCTTLIAVILGIVLVVSIKPGV TQKVGEIARTGSTPEVSTVDAMLDLIRNMFPENLVQACFQQYKTKREEVKPPSDPEMNMT EESFTAVMTTAISKNKTKEYKIVGMYSDGINVLGLIVFCLVFGLVIGKMGEKGQILVDFF NALSDATMKIVQIIMCYMPLGILFLIAGKIIEVEDWEIFRKLGLYMATVLTGLAIHSIVI LPLIYFIVVRKNPFRFAMGMAQALLTALMISSSSATLPVTFRCAEENNQVDKRITRFVLP VGATINMDGTALYEAVAAVFIAQLNDLDLGIGQIITISITATSASIGAAGVPQAGLVTMV IVLSAVGLPAEDVTLIIAVDWLLDRFRTMVNVLGDAFGTGIVEKLSKKELEQMDVSSEVN IVNPFALESTILDNEDSDTKKSYVNGGFAVDKSDTISFTQTSQF Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T46PRA | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: L-aspartic acid | Ligand Info | |||||
| Structure Description | hEAAT3-OFS-Asp | PDB:6X2Z | ||||
| Method | Electron microscopy | Resolution | 3.03 Å | Mutation | Yes | [6] |
| PDB Sequence |
KNNWVLLSTV
25 AAVVLGITTG35 VLVREHSNLS45 TLEKFYFAFP55 GEILMRMLKL65 IILPLIISSM 75 ITGVAALDSN85 VSGKIGLRAV95 VYYFCTTLIA105 VILGIVLVVS115 IKPGVTQKVG 125 STVDAMLDLI146 RNMFPENLVQ156 ACFQQYKTKR166 EEEYKIVGMY206 SDGINVLGLI 216 VFCLVFGLVI226 GKMGEKGQIL236 VDFFNALSDA246 TMKIVQIIMC256 YMPLGILFLI 266 AGKIIEVEDW276 EIFRKLGLYM286 ATVLTGLAIH296 SIVILPLIYF306 IVVRKNPFRF 316 AMGMAQALLT326 ALMISSSSAT336 LPVTFRCAEE346 NNQVDKRITR356 FVLPVGATIN 366 MDGTALYEAV376 AAVFIAQLND386 LDLGIGQIIT396 ISITATSASI406 GAAGVPQAGL 416 VTMVIVLSAV426 GLPAEDVTLI436 IAVDWLLDRF446 RTMVNVLGDA456 FGTGIVEKLS 466 KKELEQMDV
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid | Ligand Info | |||||
| Structure Description | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | PDB:7NSG | ||||
| Method | Electron microscopy | Resolution | 3.34 Å | Mutation | Yes | [7] |
| PDB Sequence |
EWKRFLKNNW
19 VLLSTVAAVV29 LGITTGVLVR39 EHRNLSTLEK49 FYFAFPGEIL59 MRMLKLIILP 69 LIISSMITGV79 AALDSNVSGK89 IGVRAVVYYF99 CTTLIAVILG109 IVLVVSIKPG 119 VTQVSTVDAM142 LDLIRNMFPE152 NLVQACFQQY162 KTKREEVKPK196 TKEYKIVGMY 206 SDGINVLGLI216 VFCLVFGLVI226 GKMGEKGQIL236 VDFFNALSDA246 TMKIVQIIMC 256 YMPLGILFLI266 AGKIIEVEDW276 EIFRKLGLYM286 ATVLTGLAIH296 SIVILPLIYF 306 IVVRKNPFRF316 AMGMAQALLT326 ALMISSSSAT336 LPVTFRCAEE346 NNQVDKRITR 356 FVLPVGATIN366 MDGTALYEAV376 AAVFIAQLND386 LDLGIGQIIT396 ISITATSASI 406 GAAGVPQAGL416 VTMVIVLSAV426 GLPAEDVTLI436 IAVDWLLDRF446 RTMVNVLGDA 456 FGTGIVEKLS466 KKELEQMDVS476
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
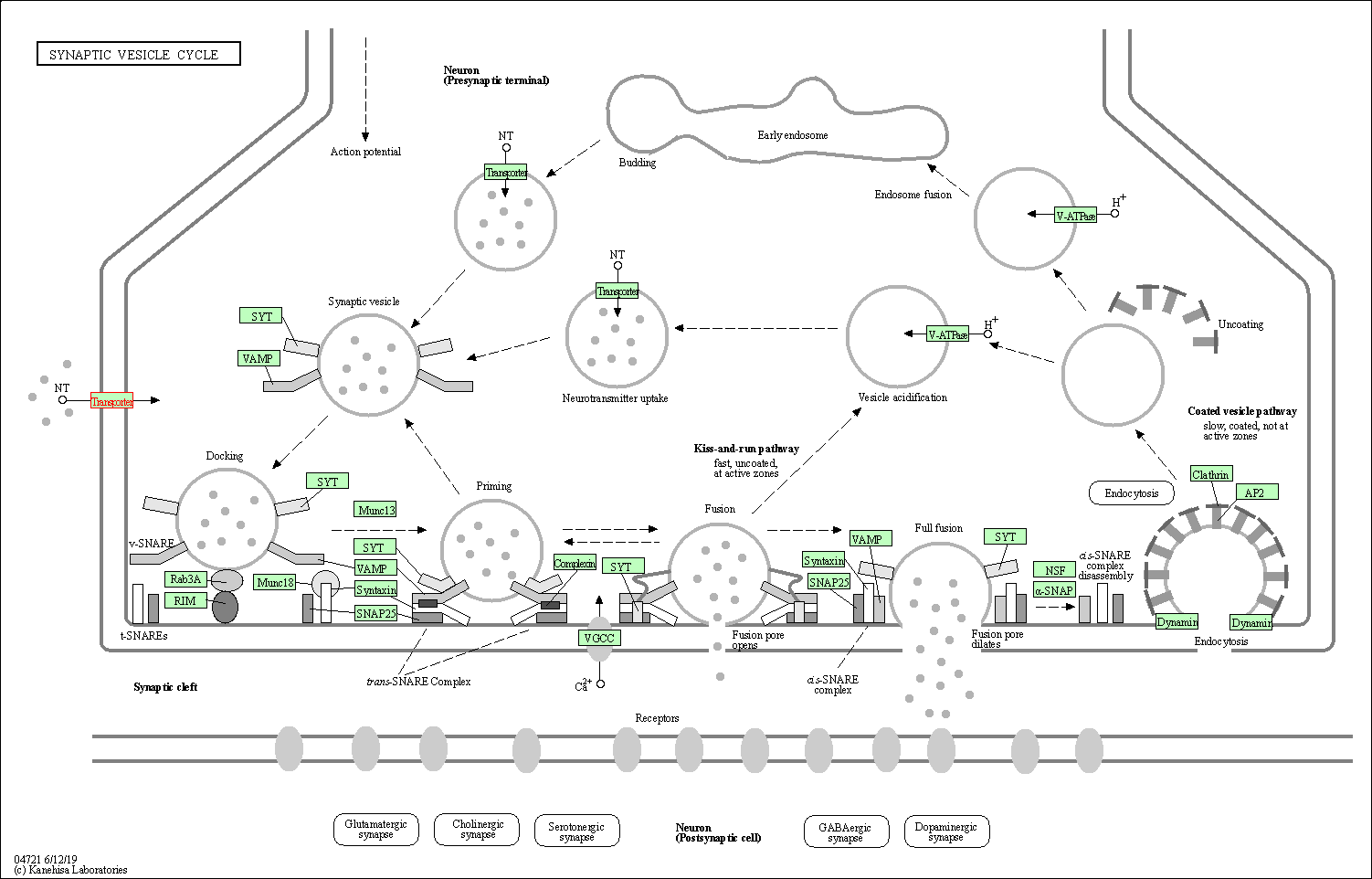
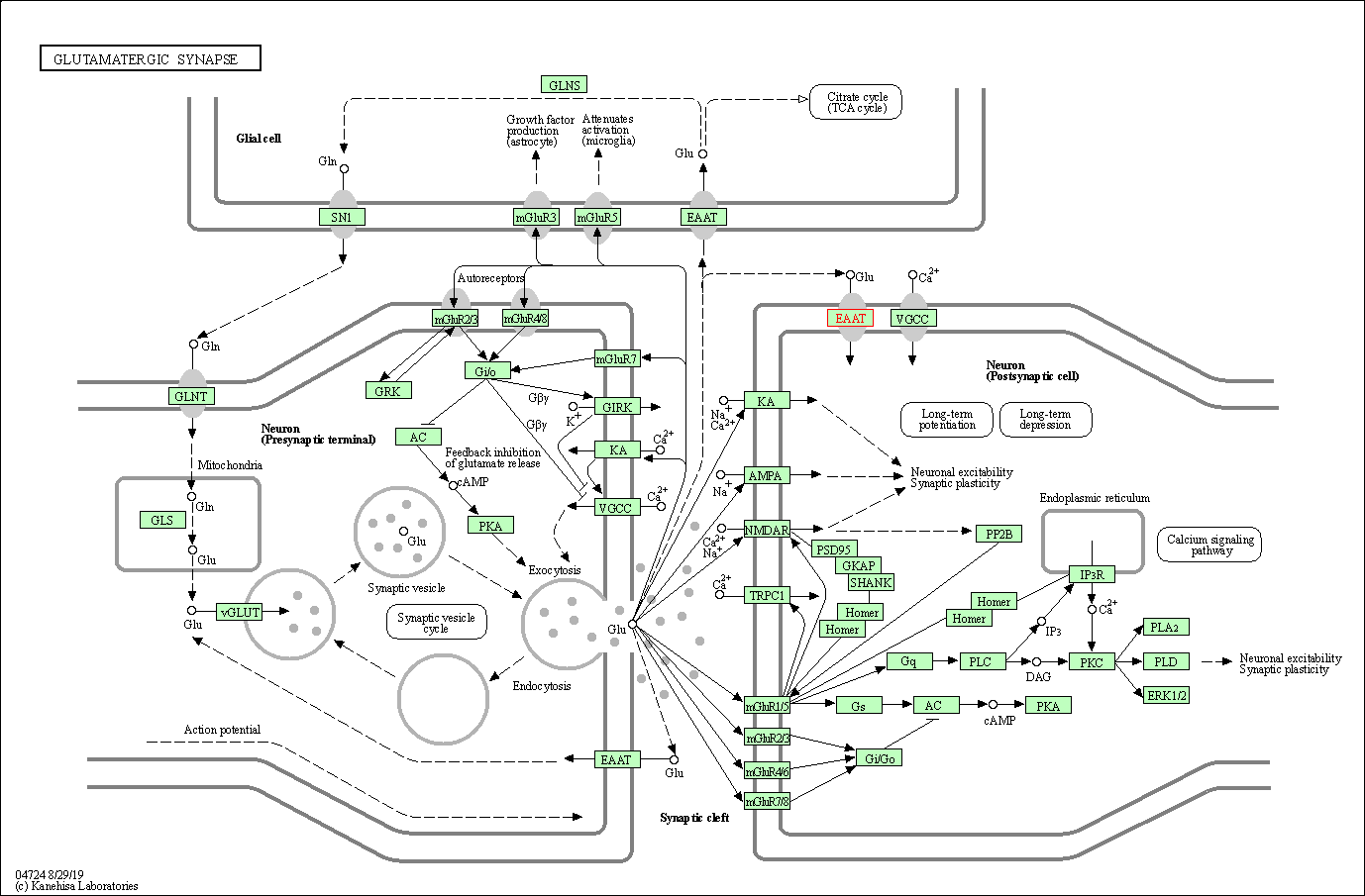
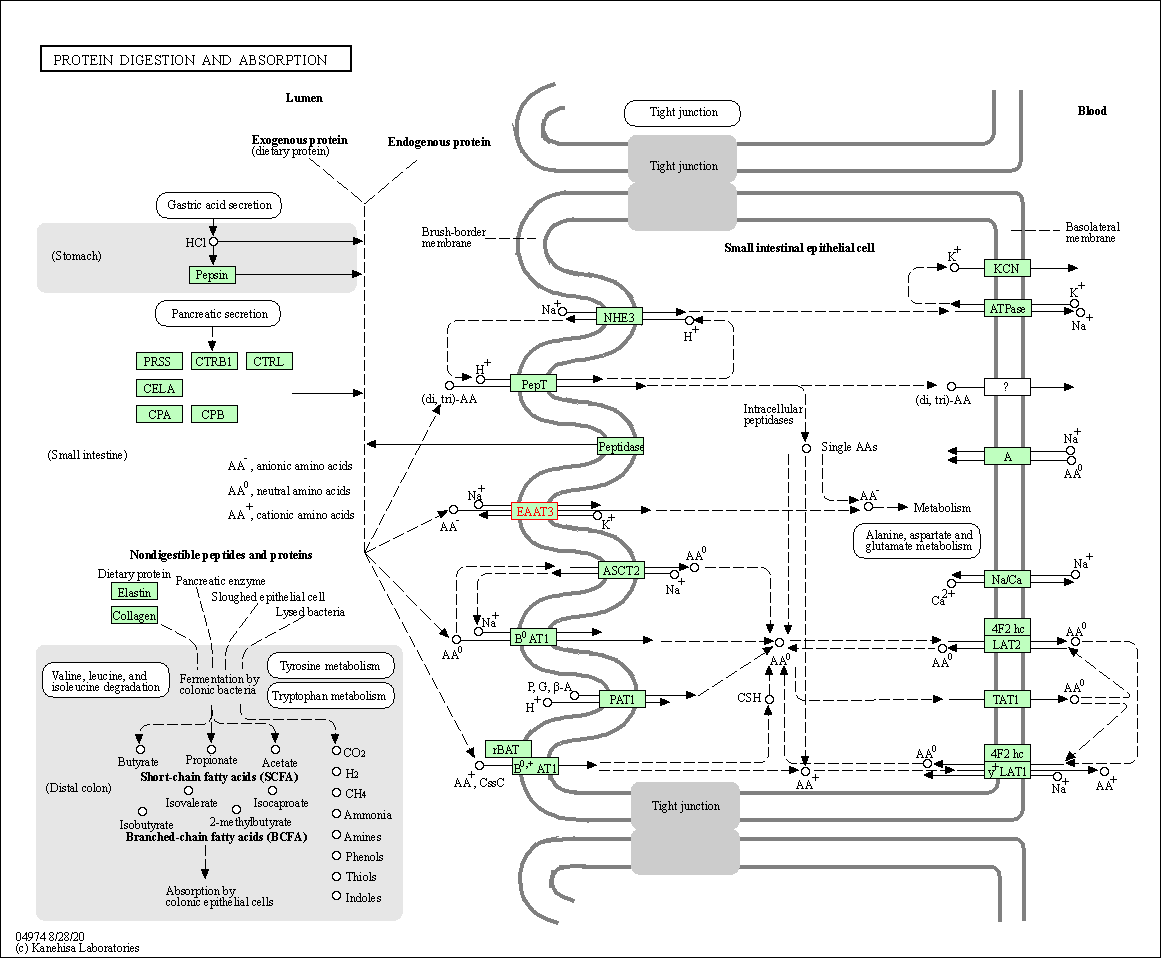
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Synaptic vesicle cycle | hsa04721 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Glutamatergic synapse | hsa04724 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Protein digestion and absorption | hsa04974 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.07E-04 | Radiality | 3.98E-03 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 1.00E+00 | Topological coefficient | . | Eccentricity | 1 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Ligands targeting the excitatory amino acid transporters (EAATs). Curr Top Med Chem. 2006;6(17):1897-906. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 870). | |||||
| REF 3 | Syntheses of optically pure beta-hydroxyaspartate derivatives as glutamate transporter blockers. Bioorg Med Chem Lett. 2000 Nov 6;10(21):2407-10. | |||||
| REF 4 | The substituted aspartate analogue L-beta-threo-benzyl-aspartate preferentially inhibits the neuronal excitatory amino acid transporter EAAT3. Neuropharmacology. 2005 Nov;49(6):850-61. | |||||
| REF 5 | Characterization of the tritium-labeled analog of L-threo-beta-benzyloxyaspartate binding to glutamate transporters. Mol Pharmacol. 2007 Jan;71(1):294-302. | |||||
| REF 6 | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport. Sci Adv. 2021 Mar 3;7(10):eabf5814. | |||||
| REF 7 | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

