Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T81240
(Former ID: TTDNR00663)
|
|||||
| Target Name |
E3 ubiquitin protein ligase Itchy (ITCH)
|
|||||
| Synonyms |
NFE2-associated polypeptide 1; NAPP1; Itch; HECT-type E3 ubiquitin transferase Itchy homolog; E3 ubiquitin-protein ligase Itchy homolog; Atrophin-1-interacting protein 4; AIP4
Click to Show/Hide
|
|||||
| Gene Name |
ITCH
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Catalyzes 'Lys-29'-, 'Lys-48'- and 'Lys-63'-linked ubiquitin conjugation. Involved in the control of inflammatory signaling pathways. Essential component of a ubiquitin-editing protein complex, comprising also TNFAIP3, TAX1BP1 and RNF11, that ensures the transient nature of inflammatory signaling pathways. Promotes the association of the complex after TNF stimulation. Once the complex is formed, TNFAIP3 deubiquitinates 'Lys-63' polyubiquitin chains on RIPK1 and catalyzes the formation of 'Lys-48'-polyubiquitin chains. This leads to RIPK1 proteasomal degradation and consequently termination of the TNF- or LPS-mediated activation of NFKB1. Ubiquitinates RIPK2 by 'Lys-63'-linked conjugation and influences NOD2-dependent signal transduction pathways. Regulates the transcriptional activity of several transcription factors, and probably plays an important role in the regulation of immune response. Ubiquitinates NFE2 by 'Lys-63' linkages and is implicated in the control of the development of hematopoietic lineages. Mediates JUN ubiquitination and degradation. Mediates JUNB ubiquitination and degradation. Critical regulator of type 2 helper T (Th2) cell cytokine production by inducing JUNB ubiquitination and degradation. Involved in the negative regulation of MAVS-dependent cellular antiviral responses. Ubiquitinates MAVS through 'Lys-48'-linked conjugation resulting in MAVS proteasomal degradation. Following ligand stimulation, regulates sorting of Wnt receptor FZD4 to the degradative endocytic pathway probably by modulating PI42KA activity. Ubiquitinates PI4K2A and negatively regulates its catalytic activity. Ubiquitinates chemokine receptor CXCR4 and regulates sorting of CXCR4 to the degradative endocytic pathway following ligand stimulation by ubiquitinating endosomal sorting complex required for transport ESCRT-0 components HGS and STAM. Targets DTX1 for lysosomal degradation and controls NOTCH1 degradation, in the absence of ligand, through 'Lys-29'-linked polyubiquitination. Ubiquitinates SNX9. Ubiquitinates MAP3K7 through 'Lys-48'-linked conjugation. Involved in the regulation of apoptosis and reactive oxygen species levels through the ubiquitination and proteasomal degradation of TXNIP. Mediates the antiapoptotic activity of epidermal growth factor through the ubiquitination and proteasomal degradation of p15 BID. Ubiquitinates BRAT1 and this ubiquitination is enhanced in the presence of NDFIP1. Acts as an E3 ubiquitin-protein ligase which accepts ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates.
Click to Show/Hide
|
|||||
| BioChemical Class |
Acyltransferase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.3.2.26
|
|||||
| Sequence |
MSDSGSQLGSMGSLTMKSQLQITVISAKLKENKKNWFGPSPYVEVTVDGQSKKTEKCNNT
NSPKWKQPLTVIVTPVSKLHFRVWSHQTLKSDVLLGTAALDIYETLKSNNMKLEEVVVTL QLGGDKEPTETIGDLSICLDGLQLESEVVTNGETTCSENGVSLCLPRLECNSAISAHCNL CLPGLSDSPISASRVAGFTGASQNDDGSRSKDETRVSTNGSDDPEDAGAGENRRVSGNNS PSLSNGGFKPSRPPRPSRPPPPTPRRPASVNGSPSATSESDGSSTGSLPPTNTNTNTSEG ATSGLIIPLTISGGSGPRPLNPVTQAPLPPGWEQRVDQHGRVYYVDHVEKRTTWDRPEPL PPGWERRVDNMGRIYYVDHFTRTTTWQRPTLESVRNYEQWQLQRSQLQGAMQQFNQRFIY GNQDLFATSQSKEFDPLGPLPPGWEKRTDSNGRVYFVNHNTRITQWEDPRSQGQLNEKPL PEGWEMRFTVDGIPYFVDHNRRTTTYIDPRTGKSALDNGPQIAYVRDFKAKVQYFRFWCQ QLAMPQHIKITVTRKTLFEDSFQQIMSFSPQDLRRRLWVIFPGEEGLDYGGVAREWFFLL SHEVLNPMYCLFEYAGKDNYCLQINPASYINPDHLKYFRFIGRFIAMALFHGKFIDTGFS LPFYKRILNKPVGLKDLESIDPEFYNSLIWVKENNIEECDLEMYFSVDKEILGEIKSHDL KPNGGNILVTEENKEEYIRMVAEWRLSRGVEEQTQAFFEGFNEILPQQYLQYFDAKELEV LLCGMQEIDLNDWQRHAIYRHYARTSKQIMWFWQFVKEIDNEKRMRLLQFVTGTCRLPVG GFADLMGSNGPQKFCIEKVGKENWLPRSHTCFNRLDLPPYKSYEQLKEKLLFAIEETEGF GQE Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
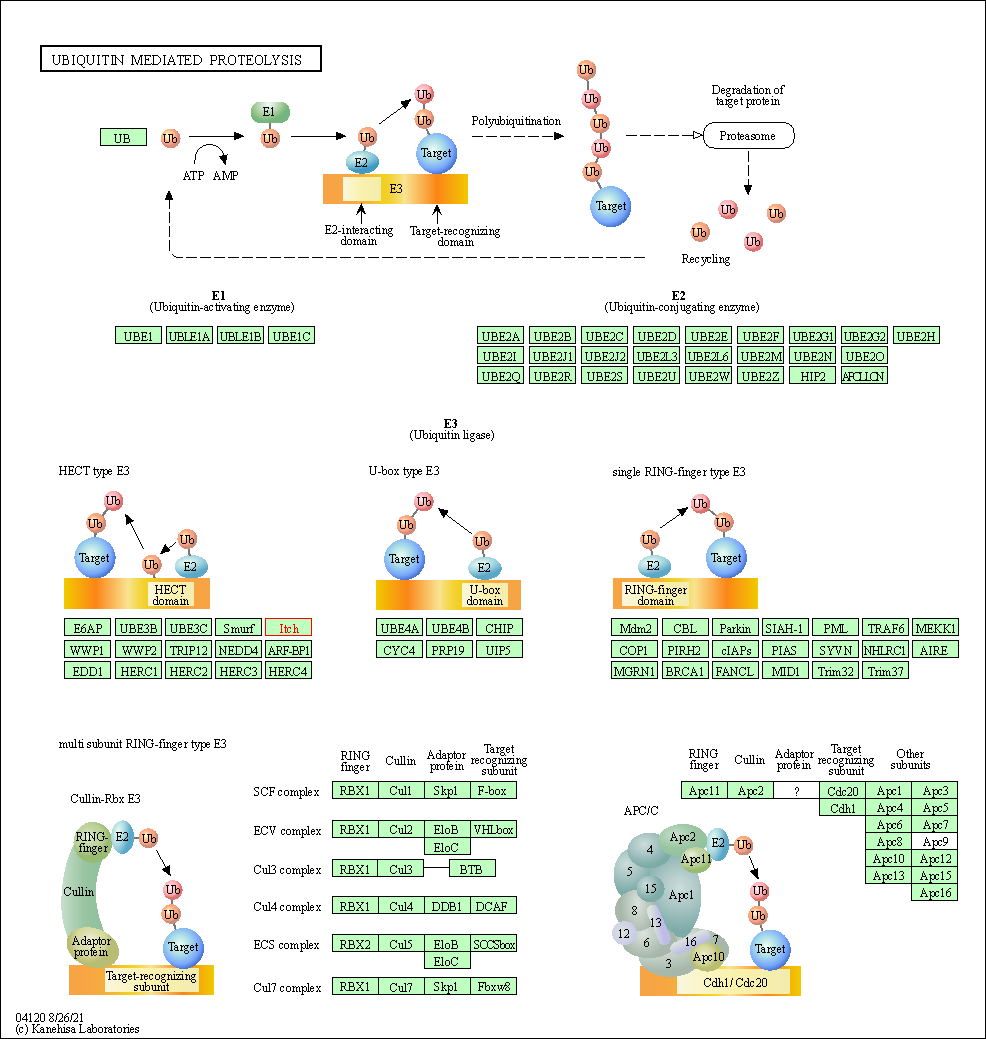
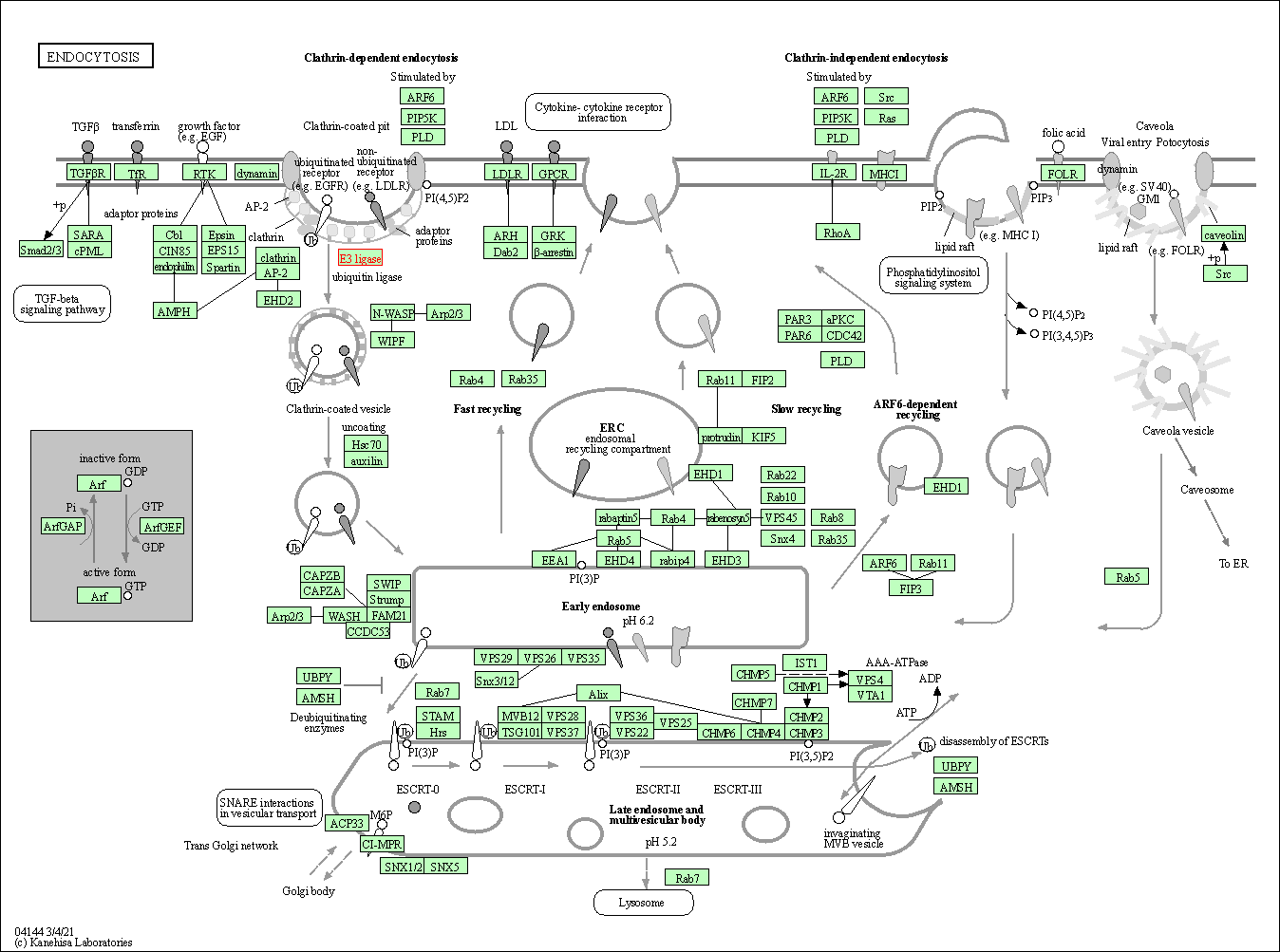
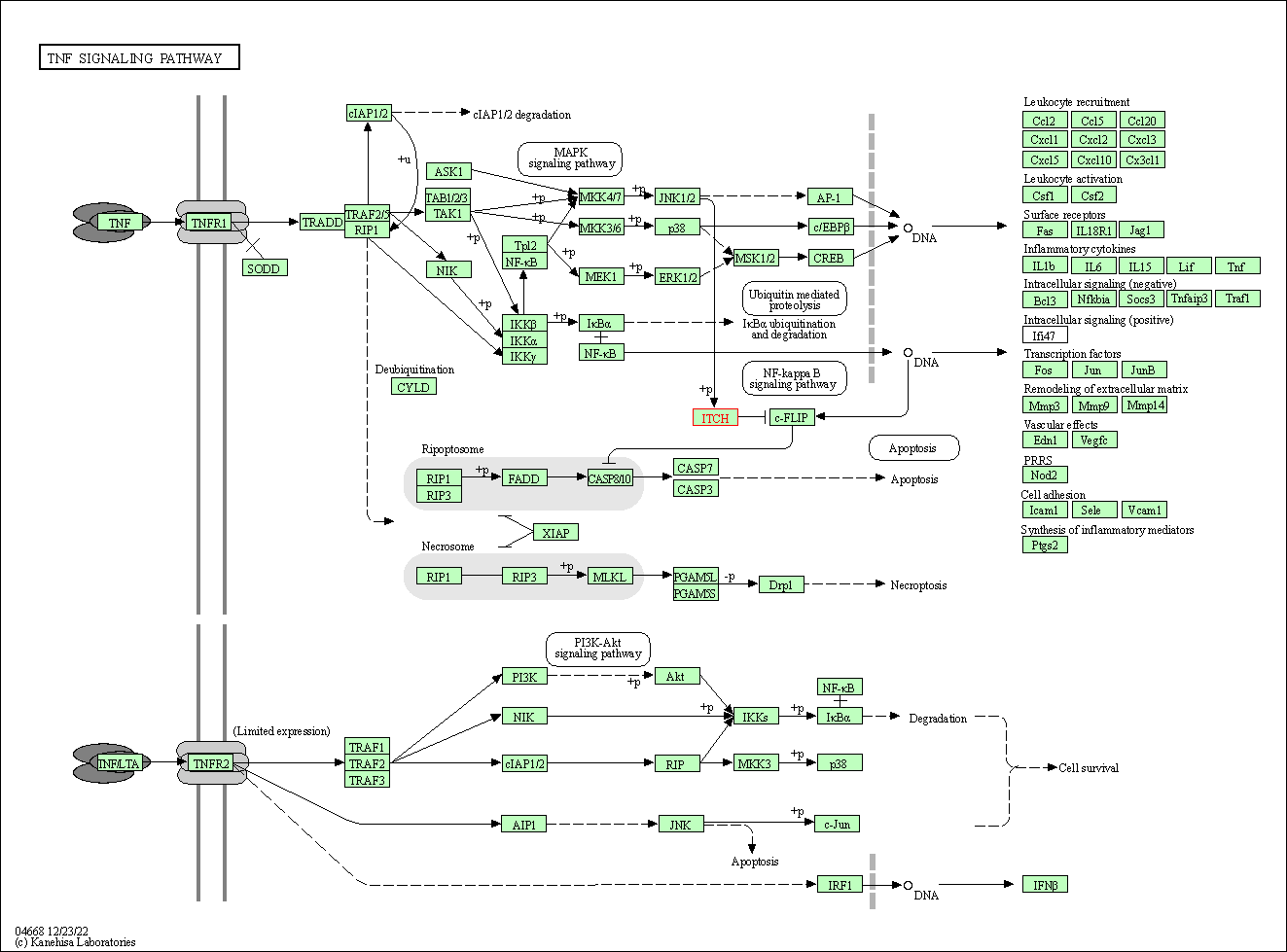
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ubiquitin mediated proteolysis | hsa04120 | Affiliated Target |

|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Endocytosis | hsa04144 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Degree | 14 | Degree centrality | 1.50E-03 | Betweenness centrality | 5.17E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.37E-01 | Radiality | 1.42E+01 | Clustering coefficient | 3.30E-02 |
| Neighborhood connectivity | 3.14E+01 | Topological coefficient | 7.99E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | The anti-HER3 (ErbB3) therapeutic antibody 9F7-F11 induces HER3 ubiquitination and degradation in tumors through JNK1/2- dependent ITCH/AIP4 activa... Oncotarget. 2016 Jun 14;7(24):37013-37029. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

