Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T84040
(Former ID: TTDS00465)
|
|||||
| Target Name |
Transformation-sensitive protein p120 (TRPA1)
|
|||||
| Synonyms |
TRPA1; Ankyrin-like with transmembrane domains protein 1; ANKTM1
Click to Show/Hide
|
|||||
| Gene Name |
TRPA1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Upper respiratory tract disorder [ICD-11: CA0Z] | |||||
| Function |
Receptor-activated non-selective cation channel involved in detection of pain and possibly also in cold perception and inner ear function. Has a central role in the pain response to endogenous inflammatory mediators and to a diverse array of volatile irritants, such as mustard oil, garlic and acrolein, an irritant from tears gas and vehicule exhaust fumes. Acts also as a ionotropic cannabinoid receptor by being activated by delta(9)- tetrahydrocannabinol (THC), the psychoactive component of marijuana. Not involved in menthol sensation. May be a component for the mechanosensitive transduction channel of hair cells in inner ear, thereby participating in the perception of sounds. Probably operated by a phosphatidylinositol second messenger system.
Click to Show/Hide
|
|||||
| BioChemical Class |
Transient receptor potential catioin channel
|
|||||
| UniProt ID | ||||||
| Sequence |
MKRSLRKMWRPGEKKEPQGVVYEDVPDDTEDFKESLKVVFEGSAYGLQNFNKQKKLKRCD
DMDTFFLHYAAAEGQIELMEKITRDSSLEVLHEMDDYGNTPLHCAVEKNQIESVKFLLSR GANPNLRNFNMMAPLHIAVQGMNNEVMKVLLEHRTIDVNLEGENGNTAVIIACTTNNSEA LQILLKKGAKPCKSNKWGCFPIHQAAFSGSKECMEIILRFGEEHGYSRQLHINFMNNGKA TPLHLAVQNGDLEMIKMCLDNGAQIDPVEKGRCTAIHFAATQGATEIVKLMISSYSGSVD IVNTTDGCHETMLHRASLFDHHELADYLISVGADINKIDSEGRSPLILATASASWNIVNL LLSKGAQVDIKDNFGRNFLHLTVQQPYGLKNLRPEFMQMQQIKELVMDEDNDGCTPLHYA CRQGGPGSVNNLLGFNVSIHSKSKDKKSPLHFAASYGRINTCQRLLQDISDTRLLNEGDL HGMTPLHLAAKNGHDKVVQLLLKKGALFLSDHNGWTALHHASMGGYTQTMKVILDTNLKC TDRLDEDGNTALHFAAREGHAKAVALLLSHNADIVLNKQQASFLHLALHNKRKEVVLTII RSKRWDECLKIFSHNSPGNKCPITEMIEYLPECMKVLLDFCMLHSTEDKSCRDYYIEYNF KYLQCPLEFTKKTPTQDVIYEPLTALNAMVQNNRIELLNHPVCKEYLLMKWLAYGFRAHM MNLGSYCLGLIPMTILVVNIKPGMAFNSTGIINETSDHSEILDTTNSYLIKTCMILVFLS SIFGYCKEAGQIFQQKRNYFMDISNVLEWIIYTTGIIFVLPLFVEIPAHLQWQCGAIAVY FYWMNFLLYLQRFENCGIFIVMLEVILKTLLRSTVVFIFLLLAFGLSFYILLNLQDPFSS PLLSIIQTFSMMLGDINYRESFLEPYLRNELAHPVLSFAQLVSFTIFVPIVLMNLLIGLA VGDIAEVQKHASLKRIAMQVELHTSLEKKLPLWFLRKVDQKSTIVYPNKPRSGGMLFHIF CFLFCTGEIRQEIPNADKSLEMEILKQKYRLKDLTFLLEKQHELIKLIIQKMEIISETED DDSHCSFQDRFKKEQMEQRNSRWNTVLRAVKAKTHHLEP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T09INZ | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Menthol | Drug Info | Approved | Throat irritation | [2], [3] | |
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | LY3526318 | Drug Info | Phase 2 | Chronic low-back pain | [4] | |
| 2 | HX-100 | Drug Info | Phase 1 | Diabetic neuropathy | [5] | |
| Mode of Action | [+] 4 Modes of Action | + | ||||
| Activator | [+] 32 Activator drugs | + | ||||
| 1 | Menthol | Drug Info | [1] | |||
| 2 | 1'-acetoxychavicol acetate | Drug Info | [7] | |||
| 3 | 1,6-hexamethylene diisocyanate | Drug Info | [8] | |||
| 4 | 2-pentenal | Drug Info | [10] | |||
| 5 | 4-oxo-nonenal | Drug Info | [12] | |||
| 6 | acetaldehyde | Drug Info | [14] | |||
| 7 | acrolein | Drug Info | [15] | |||
| 8 | artepillin C | Drug Info | [16] | |||
| 9 | benzyl bromide | Drug Info | [8] | |||
| 10 | bromoacetone | Drug Info | [8] | |||
| 11 | chlorobenzylidene malononitrile | Drug Info | [17] | |||
| 12 | chloropicrin | Drug Info | [8] | |||
| 13 | cinnamaldehyde | Drug Info | [18] | |||
| 14 | crotylaldehyde | Drug Info | [19] | |||
| 15 | dibenzoxazepine | Drug Info | [17] | |||
| 16 | dibutyl phthalate | Drug Info | [20] | |||
| 17 | Formaldehyde | Drug Info | [21] | |||
| 18 | gingerol | Drug Info | [18] | |||
| 19 | icilin | Drug Info | [22] | |||
| 20 | isovelleral | Drug Info | [23] | |||
| 21 | methyl isocyanate | Drug Info | [8] | |||
| 22 | methyl p-hydroxybenzoate | Drug Info | [24] | |||
| 23 | methyl salicylate | Drug Info | [18] | |||
| 24 | methylglyoxal | Drug Info | [25] | |||
| 25 | morphanthridine | Drug Info | [17] | |||
| 26 | MTSEA | Drug Info | [26] | |||
| 27 | NPPB | Drug Info | [27] | |||
| 28 | oleocanthal | Drug Info | [28] | |||
| 29 | omega-chloroacetophenone | Drug Info | [17] | |||
| 30 | PF-4840154 | Drug Info | [29] | |||
| 31 | prostaglandin A2 | Drug Info | [30] | |||
| 32 | super cinnamaldehyde | Drug Info | [26] | |||
| Antagonist | [+] 2 Antagonist drugs | + | ||||
| 1 | LY3526318 | Drug Info | [6] | |||
| 2 | A-967079 | Drug Info | [13] | |||
| Inhibitor | [+] 10 Inhibitor drugs | + | ||||
| 1 | HX-100 | Drug Info | [5] | |||
| 2 | 2-methyl-1-(pyridin-3-yl)pent-1-en-3-one oxime | Drug Info | [9] | |||
| 3 | 2-methyl-1-(pyridin-4-yl)pent-1-en-3-one oxime | Drug Info | [9] | |||
| 4 | 2-methyl-1-(thiophen-2-yl)pent-1-en-3-one oxime | Drug Info | [9] | |||
| 5 | 2-methyl-1-(thiophen-3-yl)pent-1-en-3-one oxime | Drug Info | [9] | |||
| 6 | 3-Methyl-4-phenylbut-3-en-2-one oxime | Drug Info | [11] | |||
| 7 | AMG-2504 | Drug Info | [9] | |||
| 8 | AMG-5445 | Drug Info | [9] | |||
| 9 | AMG-7160 | Drug Info | [9] | |||
| 10 | AMG-9090 | Drug Info | [9] | |||
| Inhibitor (gating inhibitor) | [+] 3 Inhibitor (gating inhibitor) drugs | + | ||||
| 1 | resolvin D2 | Drug Info | [31] | |||
| 2 | TCS 5861528 | Drug Info | [32] | |||
| 3 | [3H]resolvin D1 | Drug Info | [33] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Myo-inositol hexaphosphate | Ligand Info | |||||
| Structure Description | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | PDB:7OR0 | ||||
| Method | Electron microscopy | Resolution | 2.64 Å | Mutation | Yes | [34] |
| PDB Sequence |
KSPLHFAASY
456 GRINTCQRLL466 QDISDTRLLN476 EGDLHGMTPL486 HLAAKNGHDK496 VVQLLLKKGA 506 LFLSDHNGWT516 ALHHASMGGY526 TQTMKVILDT536 NLKCTDRLDE546 DGNTALHFAA 556 REGHAKAVAL566 LLSHNADIVL576 NKQQASFLHL586 ALHNKRKEVV596 LTIIRSKRWD 606 ECLKIFSHNS616 PGNKCPITEM626 IEYLPECMKV636 LLDFCMLHST646 EDKSCRDYYI 656 EYNFKYLQCP666 LEFTKKTPTQ676 DVIYEPLTAL686 NAMVQNNRIE696 LLNHPVCKEY 706 LLMKWLAYGF716 RAHMMNLGSY726 CLGLIPMTIL736 VVNIKPGMAF746 NSTGIINEEI 761 LDTTNSYLIK771 TCMILVFLSS781 IFGYCKEAGQ791 INYFMDISNV806 LEWIIYTTGI 816 IFVLPLFVEI826 PAHLQWQCGA836 IAVYFYWMNF846 LLYLQRFENC856 GIFIVMLEVI 866 LKTLLRSTVV876 FIFLLLAFGL886 SFYILLNLQD896 PFSSPLLSII906 QTFSMMLGDI 916 NYRESFLEPY926 LRNELAHPVL936 SFAQLVSFTI946 FVPIVLMNLL956 IGLAVGDIAE 966 VQKHASLKRI976 AMQVELHTSL986 EKKLPLWFLR996 KVDQKSTIVY1006 PNKPKSLEME 1043 ILKQKYRLKD1053 LTFLLEKQHE1063 LIKLIIQKME1073 IISETE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: 2-(carboxymethylamino)-2-oxoacetic acid | Ligand Info | |||||
| Structure Description | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPA1 (313-339) | PDB:6HC8 | ||||
| Method | X-ray diffraction | Resolution | 1.90 Å | Mutation | No | [35] |
| PDB Sequence |
LADYLISVGA
333 DINKID
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
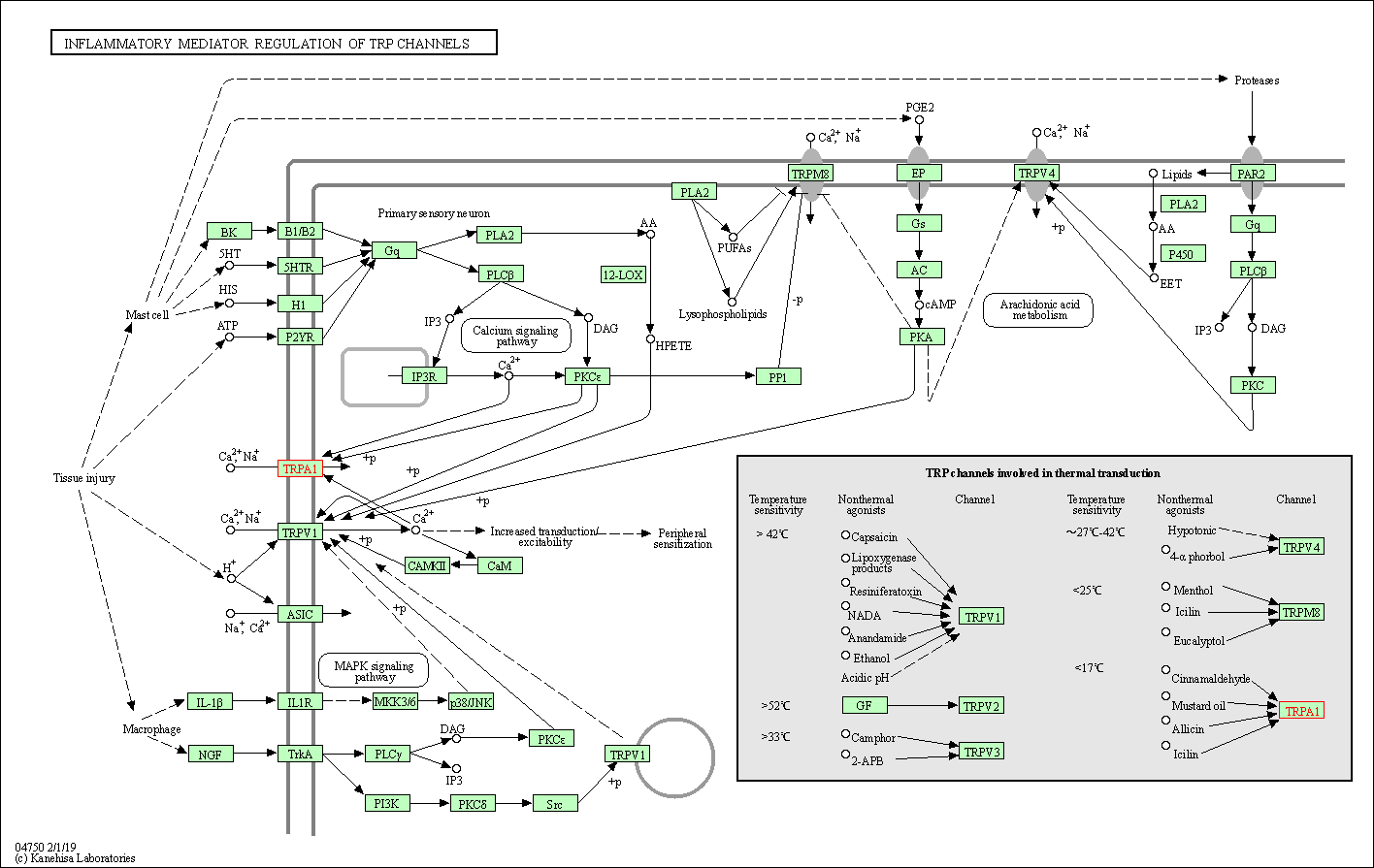
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.51E-01 | Radiality | 1.20E+01 | Clustering coefficient | 1.00E+00 |
| Neighborhood connectivity | 3.00E+00 | Topological coefficient | 7.50E-01 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Inflammatory mediator regulation of TRP channels | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | TRP channels | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | TRPV1-mediated itch in seasonal allergic rhinitis. Allergy. 2009 May;64(5):807-10. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 2471). | |||||
| REF 3 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 022029. | |||||
| REF 4 | ClinicalTrials.gov (NCT05086289) Randomized Placebo-controlled Phase 2 Clinical Trial to Evaluate LY3526318 for the Treatment of Chronic Low Back Pain. U.S.National Institutes of Health. | |||||
| REF 5 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2023. Adis Insight | |||||
| REF 7 | Galangal pungent component, 1'-acetoxychavicol acetate, activates TRPA1. Biosci Biotechnol Biochem. 2010;74(8):1694-6. | |||||
| REF 8 | Transient receptor potential ankyrin 1 antagonists block the noxious effects of toxic industrial isocyanates and tear gases. FASEB J. 2009 Apr;23(4):1102-14. | |||||
| REF 9 | Transient receptor potential ankyrin 1 (TRPA1) channel as emerging target for novel analgesics and anti-inflammatory agents. J Med Chem. 2010 Jul 22;53(14):5085-107. | |||||
| REF 10 | Transient receptor potential ankyrin 1 (TRPA1) channel as emerging target for novel analgesics and anti-inflammatory agents. J Med Chem. 2010 Jul 22;53(14):5085-107. | |||||
| REF 11 | Oxime derivatives related to AP18: Agonists and antagonists of the TRPA1 receptor. Bioorg Med Chem Lett. 2010 Jan 1;20(1):276-9. | |||||
| REF 12 | Transient receptor potential A1 is a sensory receptor for multiple products of oxidative stress. J Neurosci. 2008 Mar 5;28(10):2485-94. | |||||
| REF 13 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 485). | |||||
| REF 14 | Transient receptor potential A1 mediates acetaldehyde-evoked pain sensation. Eur J Neurosci. 2007 Nov;26(9):2516-23. | |||||
| REF 15 | TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 2006 Mar 24;124(6):1269-82. | |||||
| REF 16 | Artepillin C, a major ingredient of Brazilian propolis, induces a pungent taste by activating TRPA1 channels. PLoS One. 2012;7(11):e48072. | |||||
| REF 17 | Tear gasses CN, CR, and CS are potent activators of the human TRPA1 receptor. Toxicol Appl Pharmacol. 2008 Sep 1;231(2):150-6. | |||||
| REF 18 | Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron. 2004 Mar 25;41(6):849-57. | |||||
| REF 19 | Cigarette smoke-induced neurogenic inflammation is mediated by alpha,beta-unsaturated aldehydes and the TRPA1 receptor in rodents. J Clin Invest. 2008 Jul;118(7):2574-82. | |||||
| REF 20 | TRPA1 and TRPV1 activation is a novel adjuvant effect mechanism in contact hypersensitivity. J Neuroimmunol. 2009 Feb 15;207(1-2):66-74. | |||||
| REF 21 | An ion channel essential for sensing chemical damage. J Neurosci. 2007 Oct 17;27(42):11412-5. | |||||
| REF 22 | ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 2003 Mar 21;112(6):819-29. | |||||
| REF 23 | TRPA1 mediates the noxious effects of natural sesquiterpene deterrents. J Biol Chem. 2008 Aug 29;283(35):24136-44. | |||||
| REF 24 | Methyl p-hydroxybenzoate causes pain sensation through activation of TRPA1 channels. Br J Pharmacol. 2007 May;151(1):153-60. | |||||
| REF 25 | Inhibiting TRPA1 ion channel reduces loss of cutaneous nerve fiber function in diabetic animals: sustained activation of the TRPA1 channel contributes to the pathogenesis of peripheral diabetic neuropathy. Pharmacol Res. 2012 Jan;65(1):149-58. | |||||
| REF 26 | Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature. 2007 Feb 1;445(7127):541-5. | |||||
| REF 27 | NPPB structure-specifically activates TRPA1 channels. Biochem Pharmacol. 2010 Jul 1;80(1):113-21. | |||||
| REF 28 | Unusual pungency from extra-virgin olive oil is attributable to restricted spatial expression of the receptor of oleocanthal. J Neurosci. 2011 Jan 19;31(3):999-1009. | |||||
| REF 29 | Design and pharmacological evaluation of PF-4840154, a non-electrophilic reference agonist of the TrpA1 channel. Bioorg Med Chem Lett. 2011 Aug 15;21(16):4857-9. | |||||
| REF 30 | Cox-dependent fatty acid metabolites cause pain through activation of the irritant receptor TRPA1. Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):12045-50. | |||||
| REF 31 | Resolvin D2 is a potent endogenous inhibitor for transient receptor potential subtype V1/A1, inflammatory pain, and spinal cord synaptic plasticity in mice: distinct roles of resolvin D1, D2, and E1.J Neurosci. 2011 Dec 14;31(50):18433-8. | |||||
| REF 32 | Attenuation of mechanical hypersensitivity by an antagonist of the TRPA1 ion channel in diabetic animals. Anesthesiology. 2009 Jul;111(1):147-54. | |||||
| REF 33 | Resolvin D1 attenuates activation of sensory transient receptor potential channels leading to multiple anti-nociception. Br J Pharmacol. 2010 Oct;161(3):707-20. | |||||
| REF 34 | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60 | |||||
| REF 35 | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPA1 (313-339) | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

