Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T89361
(Former ID: TTDC00169)
|
|||||
| Target Name |
Cyclin-dependent kinase 6 (CDK6)
|
|||||
| Synonyms |
Serine/threonine-protein kinase PLSTIRE; Serine/threonine protein kinase PLSTIRE; Cell division protein kinase 6; CDKN6
Click to Show/Hide
|
|||||
| Gene Name |
CDK6
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Breast cancer [ICD-11: 2C60-2C6Y] | |||||
| 2 | Lung cancer [ICD-11: 2C25] | |||||
| 3 | Psoriasis [ICD-11: EA90] | |||||
| Function |
Phosphorylates pRB/RB1 and NPM1. Interacts with D-type G1 cyclins during interphase at G1 to form a pRB/RB1 kinase and controls the entrance into the cell cycle. Involved in initiation and maintenance of cell cycle exit during cell differentiation; prevents cell proliferation and regulates negatively cell differentiation, but is required for the proliferation of specific cell types (e. g. erythroid and hematopoietic cells). Essential for cell proliferation within the dentate gyrus of the hippocampus and the subventricular zone of the lateral ventricles. Required during thymocyte development. Promotes the production of newborn neurons, probably by modulating G1 length. Promotes, at least in astrocytes, changes in patterns of gene expression, changes in the actin cytoskeleton including loss of stress fibers, and enhanced motility during cell differentiation. Prevents myeloid differentiation by interfering with RUNX1 and reducing its transcription transactivation activity, but promotes proliferation of normal myeloid progenitors. Delays senescence. Promotes the proliferation of beta-cells in pancreatic islets of Langerhans. May play a role in the centrosome organization during the cell cycle phases. Serine/threonine-protein kinase involved in the control of the cell cycle and differentiation; promotes G1/S transition.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.22
|
|||||
| Sequence |
MEKDGLCRADQQYECVAEIGEGAYGKVFKARDLKNGGRFVALKRVRVQTGEEGMPLSTIR
EVAVLRHLETFEHPNVVRLFDVCTVSRTDRETKLTLVFEHVDQDLTTYLDKVPEPGVPTE TIKDMMFQLLRGLDFLHSHRVVHRDLKPQNILVTSSGQIKLADFGLARIYSFQMALTSVV VTLWYRAPEVLLQSSYATPVDLWSVGCIFAEMFRRKPLFRGSSDVDQLGKILDVIGLPGE EDWPRDVALPRQAFHSKSAQPIEKFVTDIDELGKDLLLKCLTFNPAKRISAYSALSHPYF QDLERCKENLDSHLPPSQNTSELNTA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T22IMH | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 5 Approved Drugs | + | ||||
| 1 | Apremilast | Drug Info | Approved | Psoriasis vulgaris | [2], [3] | |
| 2 | LY2835219 | Drug Info | Approved | Breast cancer | [4] | |
| 3 | Palbociclib | Drug Info | Approved | Breast cancer | [5] | |
| 4 | Ribociclib Succinate | Drug Info | Approved | Hormone receptor positive and HER2-negative advanced or metastatic breast cancer | [6] | |
| 5 | Trilaciclib | Drug Info | Approved | Small-cell lung cancer | [7] | |
| Clinical Trial Drug(s) | [+] 8 Clinical Trial Drugs | + | ||||
| 1 | LEE011 | Drug Info | Phase 3 | Solid tumour/cancer | [8], [9] | |
| 2 | G1T38 | Drug Info | Phase 2 | Breast cancer | [10] | |
| 3 | FCN-437 | Drug Info | Phase 1/2 | Breast cancer | [11] | |
| 4 | GLR2007 | Drug Info | Phase 1/2 | Non-small-cell lung cancer | [12] | |
| 5 | NUV-422 | Drug Info | Phase 1/2 | Malignant glioma | [13] | |
| 6 | FN-1501 | Drug Info | Phase 1 | Solid tumour/cancer | [14] | |
| 7 | G1T28-1 | Drug Info | Phase 1 | Solid tumour/cancer | [15] | |
| 8 | RGT-419B | Drug Info | Phase 1 | Breast cancer | [16] | |
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | INOC-005 | Drug Info | Preclinical | Solid tumour/cancer | [17] | |
| Discontinued Drug(s) | [+] 2 Discontinued Drugs | + | ||||
| 1 | CYC-103 | Drug Info | Terminated | Solid tumour/cancer | [18] | |
| 2 | PD-0183812 | Drug Info | Terminated | Retinoblastoma | [19] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 27 Inhibitor drugs | + | ||||
| 1 | Apremilast | Drug Info | [1] | |||
| 2 | Trilaciclib | Drug Info | [7] | |||
| 3 | G1T38 | Drug Info | [10] | |||
| 4 | FCN-437 | Drug Info | [23] | |||
| 5 | GLR2007 | Drug Info | [24] | |||
| 6 | NUV-422 | Drug Info | [13] | |||
| 7 | FN-1501 | Drug Info | [25] | |||
| 8 | RGT-419B | Drug Info | [26] | |||
| 9 | Isoquinoline 1,3-dione derivative 1 | Drug Info | [27] | |||
| 10 | Oxazolyl methylthiothiazole derivative 1 | Drug Info | [27] | |||
| 11 | PMID25726713-Compound-47 | Drug Info | [28] | |||
| 12 | PMID25726713-Compound-48 | Drug Info | [28] | |||
| 13 | PMID25726713-Compound-49 | Drug Info | [28] | |||
| 14 | PMID25726713-Compound-50 | Drug Info | [28] | |||
| 15 | PMID25726713-Compound-51 | Drug Info | [28] | |||
| 16 | INOC-005 | Drug Info | [29] | |||
| 17 | CYC-103 | Drug Info | [30] | |||
| 18 | PD-0183812 | Drug Info | [31] | |||
| 19 | 3,4-di-(4-methoxyphenyl)-1H-pyrrole-2,5-dione | Drug Info | [32] | |||
| 20 | 3,4-diphenyl-1H-pyrrole-2,5-dione | Drug Info | [32] | |||
| 21 | 3,7,3',4'-TETRAHYDROXYFLAVONE | Drug Info | [33], [34] | |||
| 22 | 3-(4-methoxyphenyl)-4-phenyl-1H-pyrrole-2,5-dione | Drug Info | [32] | |||
| 23 | APIGENIN | Drug Info | [34] | |||
| 24 | Chrysin | Drug Info | [34] | |||
| 25 | Deschloroflavopiridol | Drug Info | [31] | |||
| 26 | Fascaplysin | Drug Info | [31] | |||
| 27 | RGB-286147 | Drug Info | [35] | |||
| Modulator | [+] 5 Modulator drugs | + | ||||
| 1 | LY2835219 | Drug Info | [20] | |||
| 2 | Palbociclib | Drug Info | [5] | |||
| 3 | Ribociclib Succinate | Drug Info | [21] | |||
| 4 | LEE011 | Drug Info | [22] | |||
| 5 | G1T28-1 | Drug Info | [20] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Abemaciclib | Ligand Info | |||||
| Structure Description | The X-ray co-crystal structure of human CDK6 and Abemaciclib. | PDB:5L2S | ||||
| Method | X-ray diffraction | Resolution | 2.27 Å | Mutation | No | [36] |
| PDB Sequence |
QQYECVAEIG
20 EGAYGKVFKA30 RDLKNGGRFV40 ALKRVRVPLS57 TIREVAVLRH67 LETFEHPNVV 77 RLFDVCTKLT95 LVFEHVDQDL105 TTYLDKVPEP115 GVPTETIKDM125 MFQLLRGLDF 135 LHSHRVVHRD145 LKPQNILVTS155 SGQIKLADFG165 LAVTLWYRAP188 EVLLQSSYAT 198 PVDLWSVGCI208 FAEMFRRKPL218 FRGSSDVDQL228 GKILDVIGLP238 GEEDWPRDVA 248 LPRQAFHSKS258 AQPIEKFVTD268 IDELGKDLLL278 KCLTFNPAKR288 ISAYSALSHP 298 YFQ
|
|||||
|
|
GLU18
4.164
ILE19
2.439
GLY20
3.300
GLU21
4.733
TYR24
2.675
VAL27
3.053
ALA41
3.586
LYS43
2.907
VAL77
3.252
PHE98
3.064
GLU99
2.310
|
|||||
| Ligand Name: Palbociclib | Ligand Info | |||||
| Structure Description | The X-ray co-crystal structure of human CDK6 and Palbociclib. | PDB:5L2I | ||||
| Method | X-ray diffraction | Resolution | 2.75 Å | Mutation | No | [36] |
| PDB Sequence |
DQQYECVAEI
19 GEGAYGKVFK29 ARDLKNGGRF39 VALKRVRPLS57 TIREVAVLRH67 LETFEHPNVV 77 RLFDVCTKLT95 LVFEHVDQDL105 TTYLDKVPEP115 GVPTETIKDM125 MFQLLRGLDF 135 LHSHRVVHRD145 LKPQNILVTS155 SGQIKLADFG165 LVTLWYRAPE189 VLLQSSYATP 199 VDLWSVGCIF209 AEMFRRKPLF219 RGSSDVDQLG229 KILDVIGLPG239 EEDWPRDVAL 249 PRQAFKSAQP261 IEKFVTDIDE271 LGKDLLLKCL281 TFNPAKRISA291 YSALSHPYFQ 301
|
|||||
|
|
GLU18
4.197
ILE19
2.670
GLY20
3.575
TYR24
2.805
VAL27
2.493
ALA41
3.090
LYS43
3.867
VAL77
3.258
PHE98
2.948
GLU99
2.392
HIS100
3.550
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
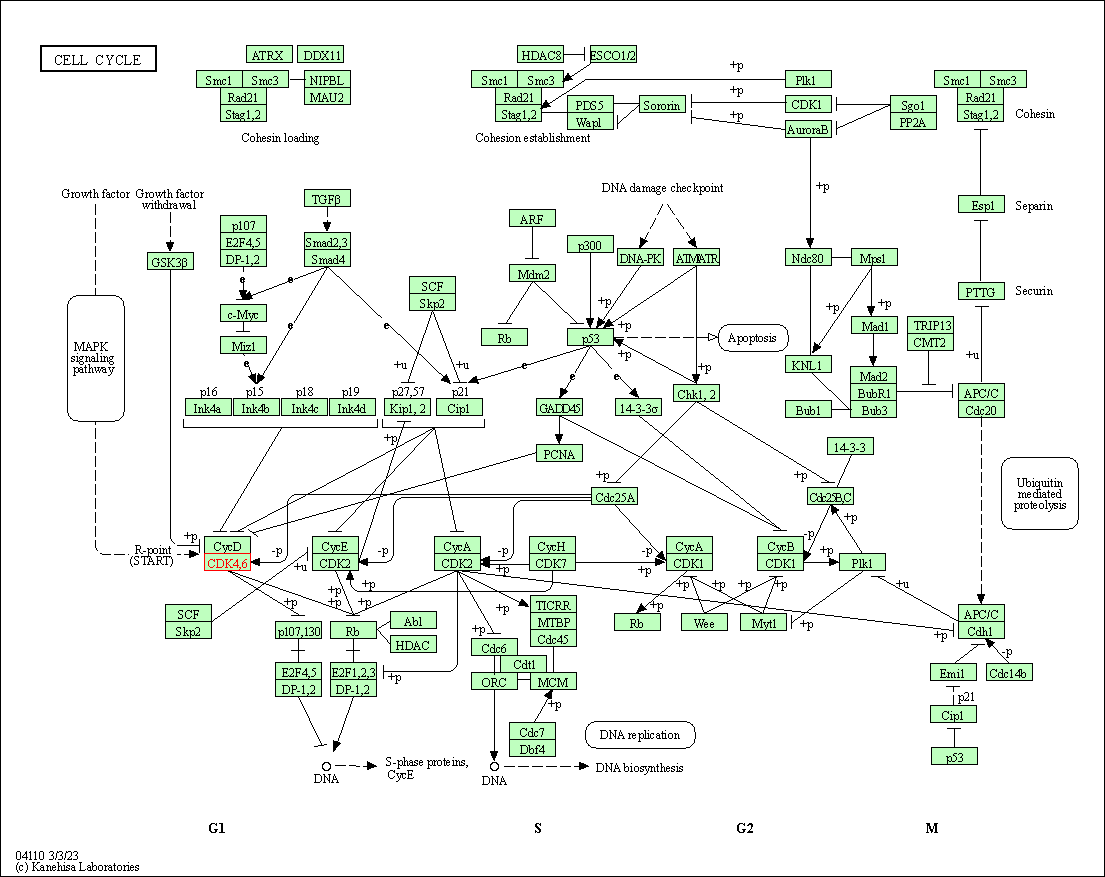
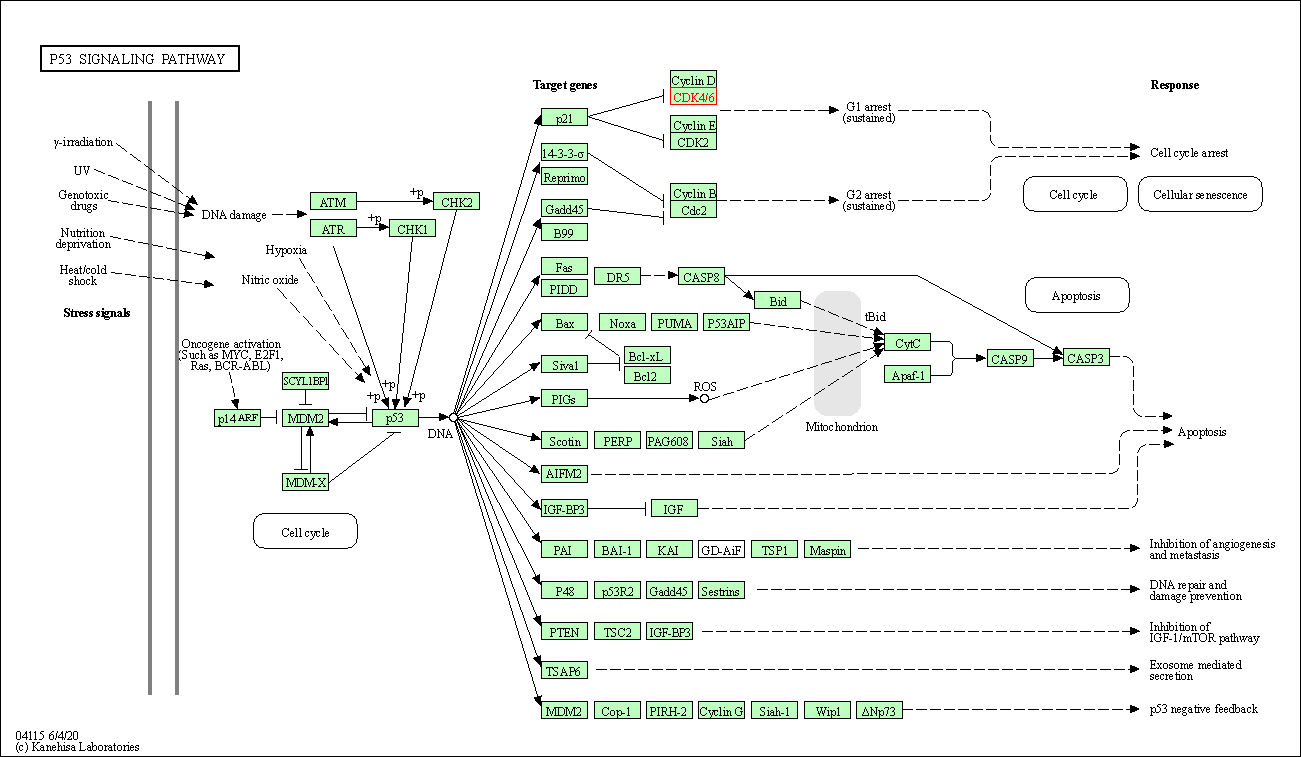
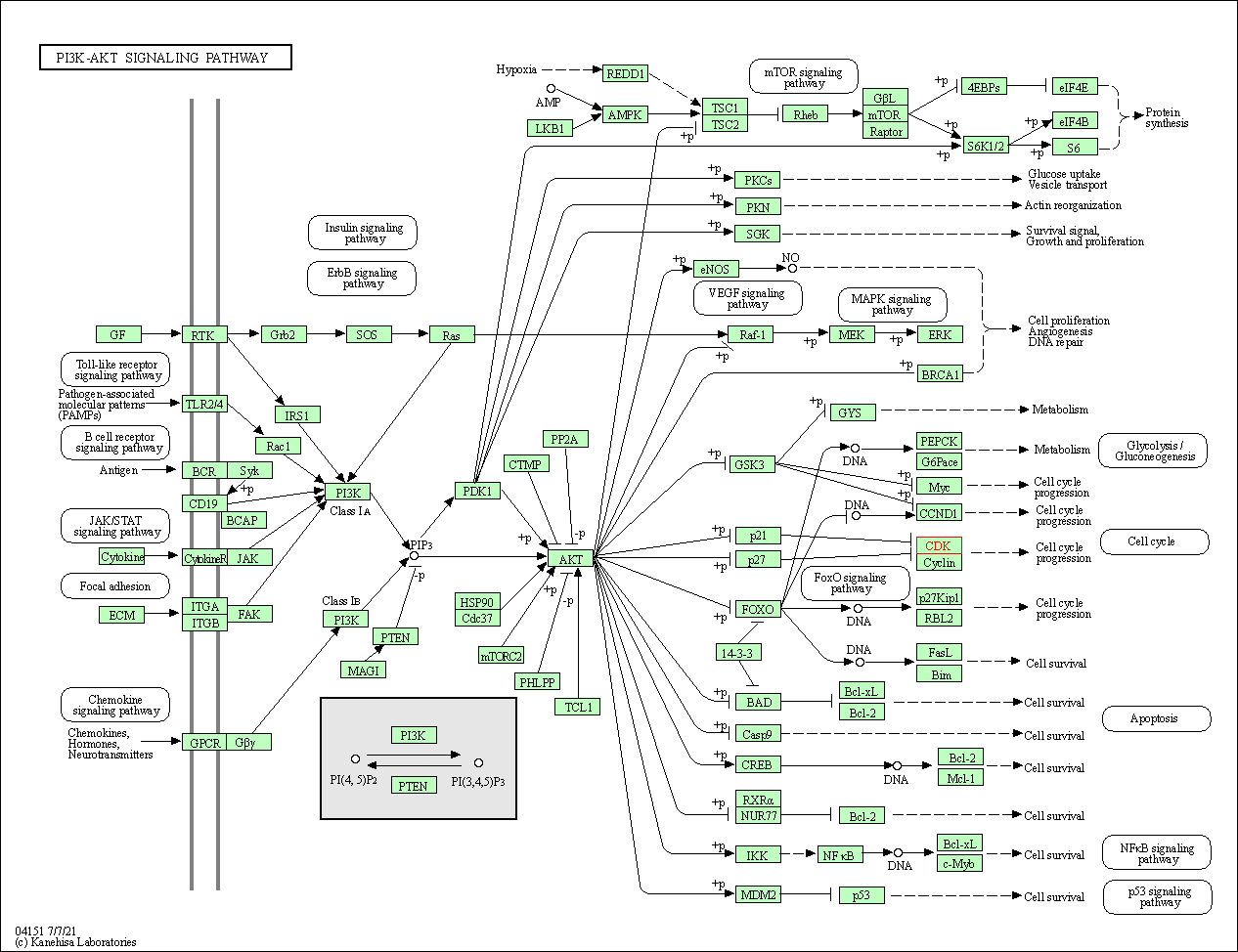
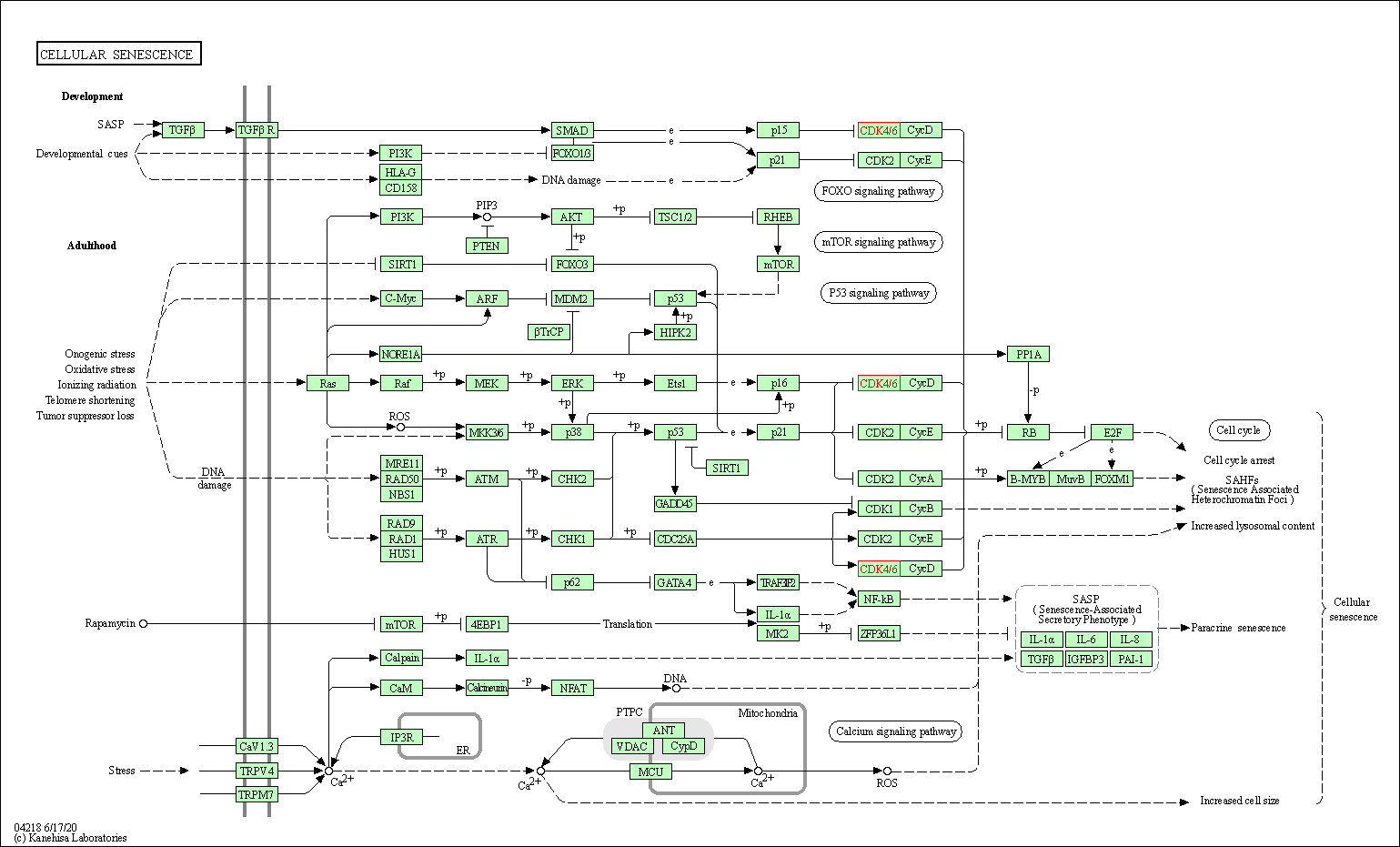
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cell cycle | hsa04110 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| p53 signaling pathway | hsa04115 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Degree | 30 | Degree centrality | 3.22E-03 | Betweenness centrality | 1.14E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.36E-01 | Radiality | 1.41E+01 | Clustering coefficient | 4.23E-01 |
| Neighborhood connectivity | 4.25E+01 | Topological coefficient | 8.76E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Agreement signed with Prostagenics to develop prostate cancer treatment. Innovate Oncology, Inc. 2005. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7372). | |||||
| REF 3 | Emerging drugs for psoriasis. Expert Opin Emerg Drugs. 2009 Mar;14(1):145-63. | |||||
| REF 4 | 2017 FDA drug approvals.Nat Rev Drug Discov. 2018 Feb;17(2):81-85. | |||||
| REF 5 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health Human Services. 2017 | |||||
| REF 6 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2018 | |||||
| REF 7 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health Human Services. 2021 | |||||
| REF 8 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7383). | |||||
| REF 9 | ClinicalTrials.gov (NCT01958021) Study of Efficacy and Safety of LEE011 in Postmenopausal Women With Advanced Breast Cancer.(MONALEESA-2). U.S. National Institutes of Health. | |||||
| REF 10 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 11 | ClinicalTrials.gov (NCT04488107) Safety, Tolerability, Pharmacokinetics and Antitumor Activity of FCN-437c. U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT04444427) Evaluation of GLR2007 for Advanced Solid Tumors. U.S. National Institutes of Health. | |||||
| REF 13 | ClinicalTrials.gov (NCT04541225) Phase 1/2 Dose Escalation, Safety, Pharmacokinetics, and Efficacy Study of NUV-422 in Adults With Recurrent or Refractory High-grade Gliomas and Solid Tumors. U.S.National Institutes of Health. | |||||
| REF 14 | ClinicalTrials.gov (NCT03690154) A Phase 1 Study to Evaluate FN-1501 Monotherapy in Patients With Advanced Solid Tumors and R/R AML. U.S. National Institutes of Health. | |||||
| REF 15 | ClinicalTrials.gov (NCT02243150) Safety, Pharmacokinetic and Pharmacodynamic Study of the CDK 4/6 Inhibitor G1T28-1. U.S. National Institutes of Health. | |||||
| REF 16 | ClinicalTrials.gov (NCT05304962) First-in-Human, Escalating Oral Dose Study of RGT-419B Given Alone and With Endocrine Therapy in Subjects With Hormone Receptor Positive, Human Epidermal Growth Factor Receptor 2 Negative Advanced/Metastatic Breast Cancer. U.S.National Institutes of Health. | |||||
| REF 17 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800022337) | |||||
| REF 18 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800015455) | |||||
| REF 19 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800014130) | |||||
| REF 20 | Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41. | |||||
| REF 21 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. | |||||
| REF 22 | Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013 Nov 15;19(22):6173-82. | |||||
| REF 23 | Clinical pipeline report, company report or official report of Fochon Pharmaceuticals. | |||||
| REF 24 | Clinical pipeline report, company report or official report of Gan & Lee Pharmaceuticals. | |||||
| REF 25 | Discovery of 4-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-N-(4-((4-methylpiperazin-1-yl)methyl)phenyl)-1H-pyrazole-3-carboxamide (FN-1501), an FLT3- and CDK-Kinase Inhibitor with Potentially High Efficiency against Acute Myelocytic Leukemia. J Med Chem. 2018 Feb 22;61(4):1499-1518. | |||||
| REF 26 | Clinical pipeline report, company report or official report of Regor Therapeutics | |||||
| REF 27 | Cyclin-dependent kinase inhibitors for cancer therapy: a patent review (2009 - 2014).Expert Opin Ther Pat. 2015;25(9):953-70. | |||||
| REF 28 | Hedgehog inhibitors: a patent review (2013 - present).Expert Opin Ther Pat. 2015 May;25(5):549-65. | |||||
| REF 29 | What are next generation innovative therapeutic targets. J Pharmacol Exp Ther. 2009 Jul;330(1):304-15. | |||||
| REF 30 | WO patent application no. 2007,0898,78, Sutures and anti-scarring agents. | |||||
| REF 31 | Pharmacological inhibitors of cyclin-dependent kinases. Trends Pharmacol Sci. 2002 Sep;23(9):417-25. | |||||
| REF 32 | Design, synthesis, and biological evaluation of 3,4-diarylmaleimides as angiogenesis inhibitors. J Med Chem. 2006 Feb 23;49(4):1271-81. | |||||
| REF 33 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 34 | Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin. J Med Chem. 2005 Feb 10;48(3):737-43. | |||||
| REF 35 | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20523-8. | |||||
| REF 36 | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance. Mol Cancer Ther. 2016 Oct;15(10):2273-2281. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

