Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T97765
(Former ID: TTDR00921)
|
|||||
| Target Name |
Macrophage inflammatory protein 1-alpha (CCL3)
|
|||||
| Synonyms |
Tonsillar lymphocyte LD78 alpha protein; Small-inducible cytokine A3; SIS-beta; SCYA3; PAT 464.1; Macrophage Inflammatory Protein-1alpha Nuclear Protein; MNP; MIP1A; MIP-1-alpha; G0S19-1 protein; G0S19-1; G0/G1 switch regulatory protein 19-1; C-C motif chemokine 3
Click to Show/Hide
|
|||||
| Gene Name |
CCL3
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Transplanted organ/tissue [ICD-11: QB63] | |||||
| Function |
Binds to CCR1, CCR4 and CCR5. One of the major HIV-suppressive factors produced by CD8+ T-cells. Recombinant MIP-1-alpha induces a dose-dependent inhibition of different strains of HIV-1, HIV-2, and simian immunodeficiency virus (SIV). Monokine with inflammatory and chemokinetic properties.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cytokine: CC chemokine
|
|||||
| UniProt ID | ||||||
| Sequence |
MQVSTAALAVLLCTMALCNQFSASLAADTPTACCFSYTSRQIPQNFIADYFETSSQCSKP
GVIFLTKRSRQVCADPSEEWVQKYVSDLELSA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T92WFW | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Nagrestipen | Drug Info | Phase 1 | Bone marrow transplantation | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Nagrestipen | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Beta-D-Glucose | Ligand Info | |||||
| Structure Description | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | PDB:5D65 | ||||
| Method | X-ray diffraction | Resolution | 3.10 Å | Mutation | No | [2] |
| PDB Sequence |
SLAADTPTAC
11 CFSYTSRQIP21 QNFIADYFET31 SSQCSKPGVI41 FLTKRSRQVC51 ADPSEEWVQK 61 YVSDLELS
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
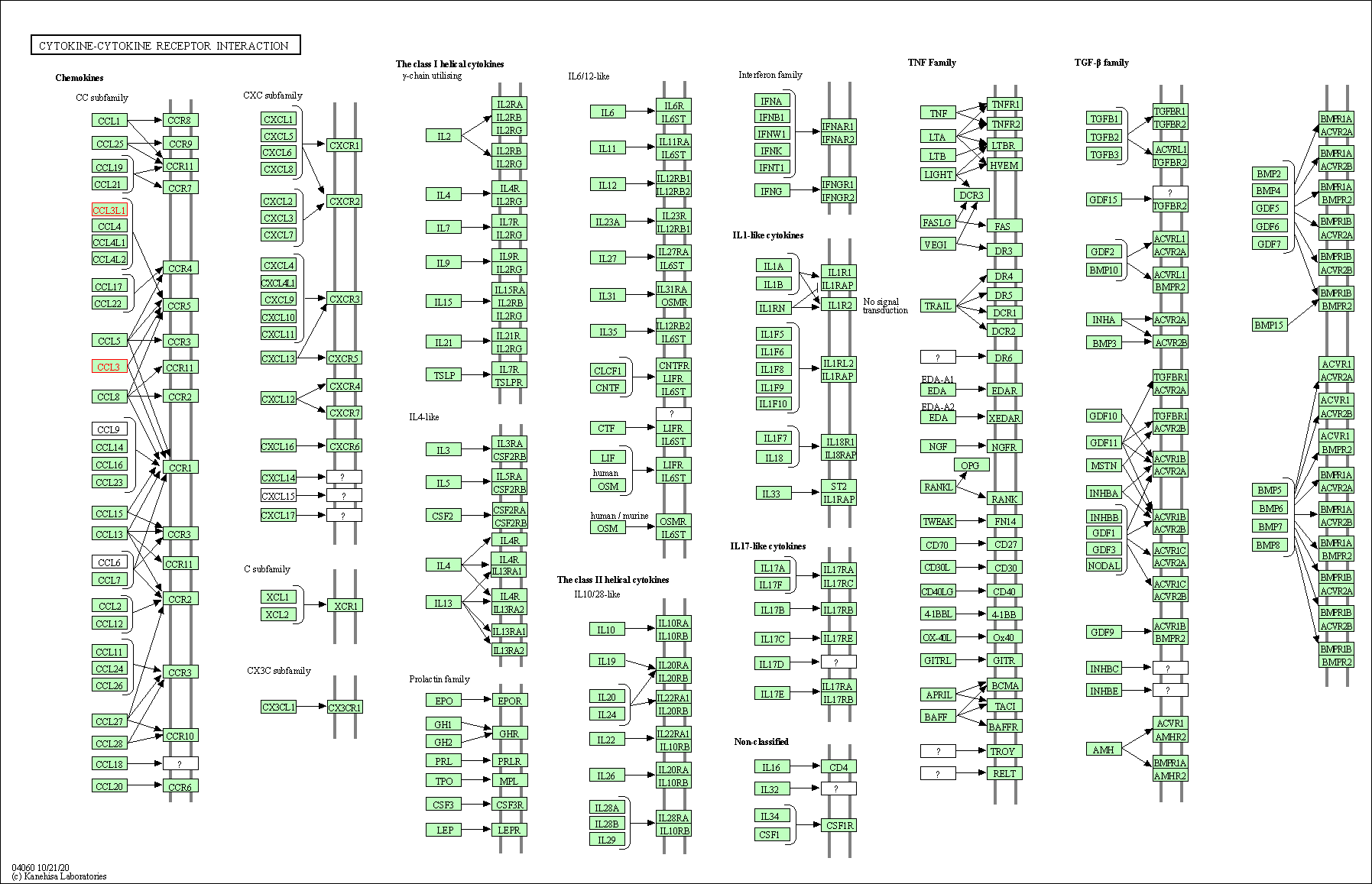
| Cytokine-cytokine receptor interaction | hsa04060 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
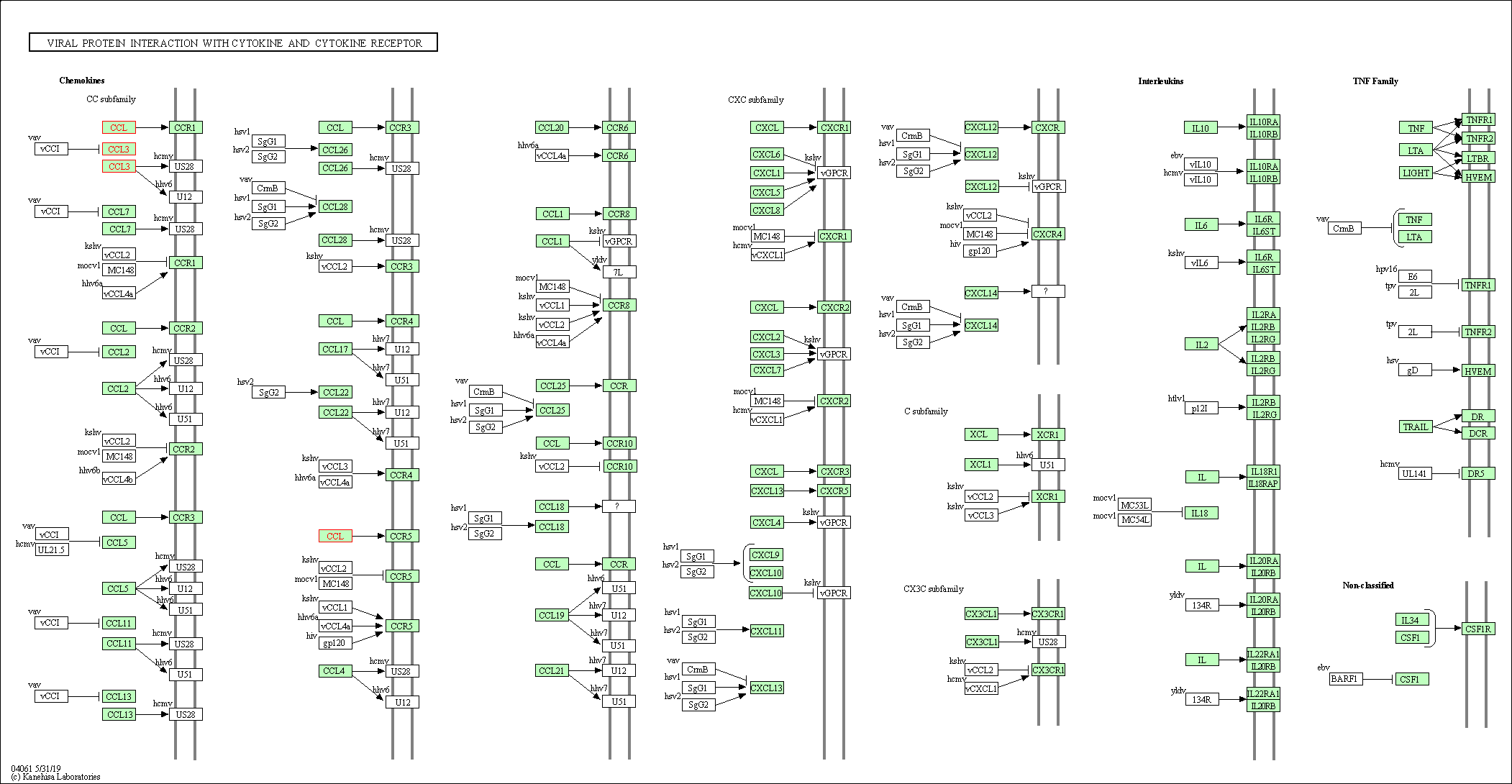
| Viral protein interaction with cytokine and cytokine receptor | hsa04061 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
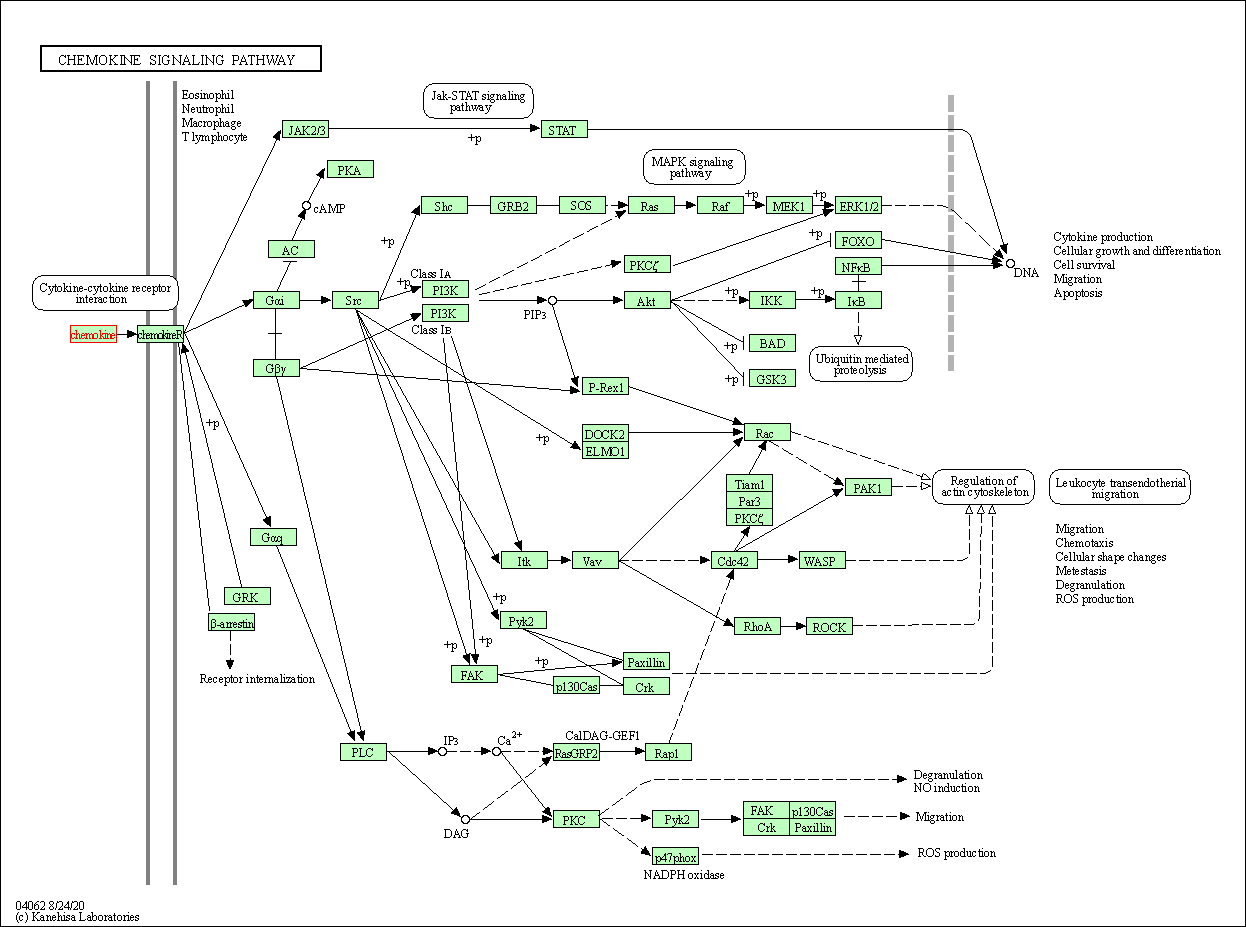
| Chemokine signaling pathway | hsa04062 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
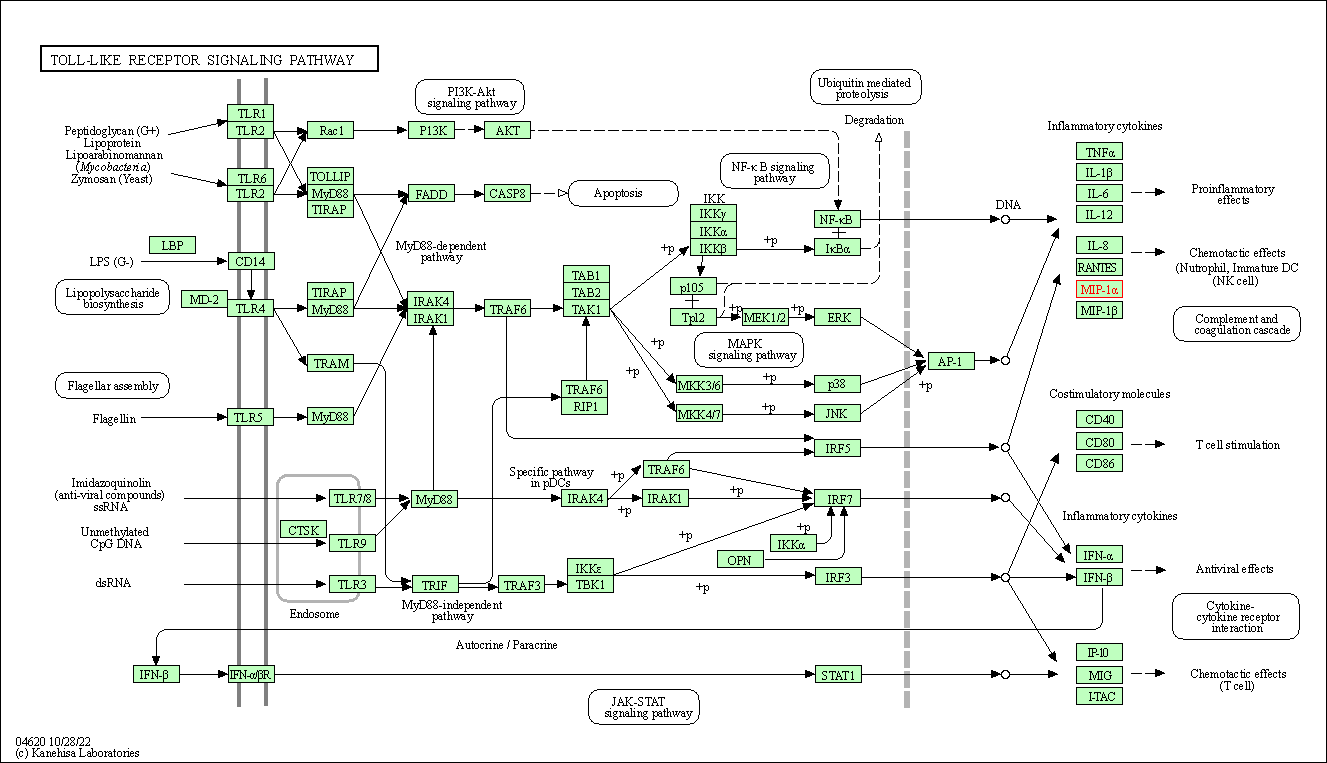
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 20 | Degree centrality | 2.15E-03 | Betweenness centrality | 1.13E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.16E-01 | Radiality | 1.38E+01 | Clustering coefficient | 5.42E-01 |
| Neighborhood connectivity | 2.71E+01 | Topological coefficient | 1.38E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 6 KEGG Pathways | + | ||||
| 1 | Cytokine-cytokine receptor interaction | |||||
| 2 | Chemokine signaling pathway | |||||
| 3 | Toll-like receptor signaling pathway | |||||
| 4 | Salmonella infection | |||||
| 5 | Chagas disease (American trypanosomiasis) | |||||
| 6 | Rheumatoid arthritis | |||||
| NetPath Pathway | [+] 6 NetPath Pathways | + | ||||
| 1 | IL9 Signaling Pathway | |||||
| 2 | IL5 Signaling Pathway | |||||
| 3 | IL2 Signaling Pathway | |||||
| 4 | TWEAK Signaling Pathway | |||||
| 5 | RANKL Signaling Pathway | |||||
| 6 | TCR Signaling Pathway | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Inflammation mediated by chemokine and cytokine signaling pathway | |||||
| PID Pathway | [+] 1 PID Pathways | + | ||||
| 1 | IL12-mediated signaling events | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Chemokine receptors bind chemokines | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Toll-like receptor signaling pathway | |||||
| 2 | Senescence and Autophagy in Cancer | |||||
| 3 | GPCR ligand binding | |||||
| 4 | Regulation of toll-like receptor signaling pathway | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Macrophage inflammatory protein derivative ECI301 enhances the alarmin-associated abscopal benefits of tumor radiotherapy. Cancer Res. 2014 Sep 15;74(18):5070-8. | |||||
| REF 2 | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3. Proc Natl Acad Sci U S A. 2016 May 3;113(18):5000-5. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

