Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T01003
(Former ID: TTDI03172)
|
|||||
| Target Name |
Endoplasmic reticulum to nucleus signaling 1 (ERN1)
|
|||||
| Synonyms |
Serine/threonine-protein kinase/endoribonuclease IRE1; Ire1-alpha; IRE1a; IRE1; Inositol-requiring protein 1; hIRE1p; Endoplasmic reticulum-to-nucleus signaling 1
Click to Show/Hide
|
|||||
| Gene Name |
ERN1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Breast cancer [ICD-11: 2C60-2C6Y] | |||||
| Function |
In unstressed cells, the endoplasmic reticulum luminal domain is maintained in its inactive monomeric state by binding to the endoplasmic reticulum chaperone HSPA5/BiP. Accumulation of misfolded protein in the endoplasmic reticulum causes release of HSPA5/BiP, allowing the luminal domain to homodimerize, promoting autophosphorylation of the kinase domain and subsequent activation of the endoribonuclease activity. The endoribonuclease activity is specific for XBP1 mRNA and excises 26 nucleotides from XBP1 mRNA. The resulting spliced transcript of XBP1 encodes a transcriptional activator protein that up-regulates expression of UPR target genes. Acts as an upstream signal for ER stress-induced GORASP2-mediated unconventional (ER/Golgi-independent) trafficking of CFTR to cell membrane by modulating the expression and localization of SEC16A. Serine/threonine-protein kinase and endoribonuclease that acts as a key sensor for the endoplasmic reticulum unfolded protein response (UPR).
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| Sequence |
MPARRLLLLLTLLLPGLGIFGSTSTVTLPETLLFVSTLDGSLHAVSKRTGSIKWTLKEDP
VLQVPTHVEEPAFLPDPNDGSLYTLGSKNNEGLTKLPFTIPELVQASPCRSSDGILYMGK KQDIWYVIDLLTGEKQQTLSSAFADSLCPSTSLLYLGRTEYTITMYDTKTRELRWNATYF DYAASLPEDDVDYKMSHFVSNGDGLVVTVDSESGDVLWIQNYASPVVAFYVWQREGLRKV MHINVAVETLRYLTFMSGEVGRITKWKYPFPKETEAKSKLTPTLYVGKYSTSLYASPSMV HEGVAVVPRGSTLPLLEGPQTDGVTIGDKGECVITPSTDVKFDPGLKSKNKLNYLRNYWL LIGHHETPLSASTKMLERFPNNLPKHRENVIPADSEKKSFEEVINLVDQTSENAPTTVSR DVEEKPAHAPARPEAPVDSMLKDMATIILSTFLLIGWVAFIITYPLSMHQQQQLQHQQFQ KELEKIQLLQQQQQQLPFHPPGDTAQDGELLDTSGPYSESSGTSSPSTSPRASNHSLCSG SSASKAGSSPSLEQDDGDEETSVVIVGKISFCPKDVLGHGAEGTIVYRGMFDNRDVAVKR ILPECFSFADREVQLLRESDEHPNVIRYFCTEKDRQFQYIAIELCAATLQEYVEQKDFAH LGLEPITLLQQTTSGLAHLHSLNIVHRDLKPHNILISMPNAHGKIKAMISDFGLCKKLAV GRHSFSRRSGVPGTEGWIAPEMLSEDCKENPTYTVDIFSAGCVFYYVISEGSHPFGKSLQ RQANILLGACSLDCLHPEKHEDVIARELIEKMIAMDPQKRPSAKHVLKHPFFWSLEKQLQ FFQDVSDRIEKESLDGPIVKQLERGGRAVVKMDWRENITVPLQTDLRKFRTYKGGSVRDL LRAMRNKKHHYRELPAEVRETLGSLPDDFVCYFTSRFPHLLAHTYRAMELCSHERLFQPY YFHEPPEPQPPVTPDAL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T93PLN | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | ORIN1001 | Drug Info | Phase 1/2 | Metastatic breast cancer | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 14 Inhibitor drugs | + | ||||
| 1 | ORIN1001 | Drug Info | [3] | |||
| 2 | 879325-11-8 | Drug Info | [4] | |||
| 3 | AC1LE05K | Drug Info | [5] | |||
| 4 | BB 0223767 | Drug Info | [6] | |||
| 5 | T8361 | Drug Info | [6] | |||
| 6 | US8614253, 29-1 | Drug Info | [7] | |||
| 7 | US8614253, 29-19 | Drug Info | [8] | |||
| 8 | US8614253, 32-4 | Drug Info | [8] | |||
| 9 | US8614253, 32-5 | Drug Info | [6] | |||
| 10 | US9040714, 155 | Drug Info | [4] | |||
| 11 | US9040714, 7 | Drug Info | [4] | |||
| 12 | US9241942, 32-12 | Drug Info | [8] | |||
| 13 | APY29 | Drug Info | [9] | |||
| 14 | PMID24749861C34 | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: L-serine-O-phosphate | Ligand Info | |||||
| Structure Description | Crystal structure of IRE1 in complex with G-3053 | PDB:6XDD | ||||
| Method | X-ray diffraction | Resolution | 2.40 Å | Mutation | No | [10] |
| PDB Sequence |
ETSVVIVGKI
569 SFCPKDVLGH579 GAEGTIVYRG589 MFDNRDVAVK599 RILPECFSFA609 DREVQLLRES 619 DEHPNVIRYF629 CTEKDRQFQY639 IAIELCAATL649 QEYVEQKDFA659 HLGLEPITLL 669 QQTTSGLAHL679 HSLNIVHRDL689 KPHNILISMP699 NAHGKIKAMI709 SDFGLCKKLA 719 VGRHFRRGVP732 GTEGWIAPEM742 LSEDCKENPT752 YTVDIFSAGC762 VFYYVISEGS 772 HPFGKSLQRQ782 ANILLGACSL792 DCLHPEKHED802 VIARELIEKM812 IAMDPQKRPS 822 AKHVLKHPFF832 WSLEKQLQFF842 QDVSDRIEKE852 SLDGPIVKQL862 ERGGRAVVKM 872 DWRENITVPL882 QTDLRKFRTY892 KGGSVRDLLR902 AMRNKKHHYR912 ELPAEVRETL 922 GSLPDDFVCY932 FTSRFPHLLA942 HTYRAMELCS952 HERLFQPYYF962 H |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: adenosine diphosphate | Ligand Info | |||||
| Structure Description | Crystal structure of the Human kinase and RNase domains in complex with ADP | PDB:3P23 | ||||
| Method | X-ray diffraction | Resolution | 2.70 Å | Mutation | No | [11] |
| PDB Sequence |
VVIVGKISFC
572 PKDVLGHGAE582 GTIVYRGMFD592 NRDVAVKRIL602 PECFSFADRE612 VQLLRESDEH 622 PNVIRYFCTE632 KDRQFQYIAI642 ELCAATLQEY652 VEQKDFAHLG662 LEPITLLQQT 672 TSGLAHLHSL682 NIVHRDLKPH692 NILISMPNAH702 GKIKAMISDF712 GLCKKLVPGT 734 EGWIAPEMLS744 EDNPTYTVDI757 FSAGCVFYYV767 ISEGSHPFGK777 SLQRQANILL 787 GACSLDCLHP797 EKHEDVIARE807 LIEKMIAMDP817 QKRPSAKHVL827 KHPFFWSLEK 837 QLQFFQDVSD847 RIEKESLDGP857 IVKQLERGGR867 AVVKMDWREN877 ITVPLQTDLR 887 KFRTYKGGSV897 RDLLRAMRNK907 KHHYRELPAE917 VRETLGTLPD927 DFVCYFTSRF 937 PHLLAHTYRA947 MELCSHERLF957 QPYYFH
|
|||||
|
|
LEU577
3.951
GLY578
3.837
HIS579
3.355
GLY580
3.030
ALA581
4.456
THR584
3.329
VAL586
3.709
ALA597
3.487
LYS599
2.690
ILE626
4.702
ILE642
3.617
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
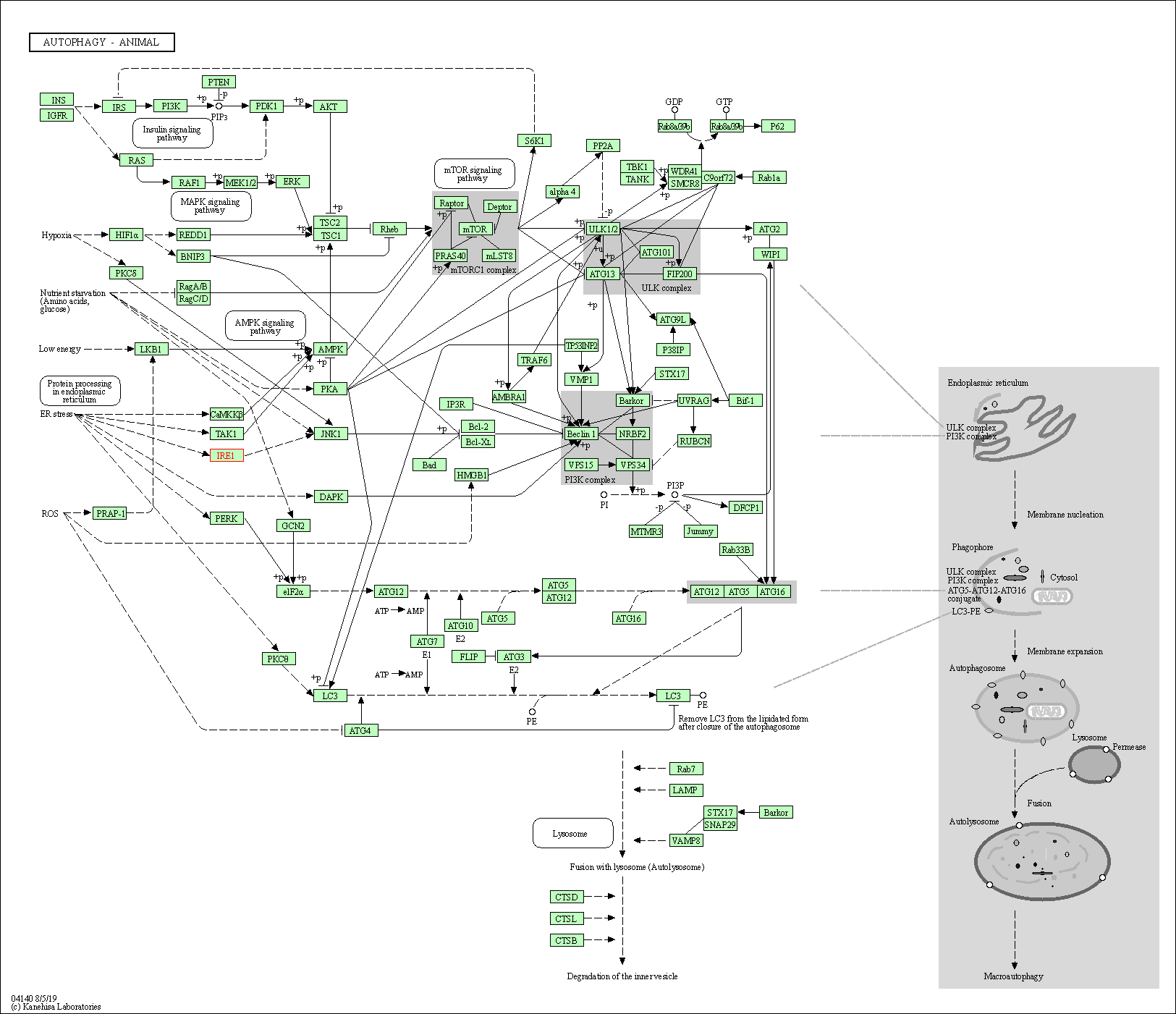
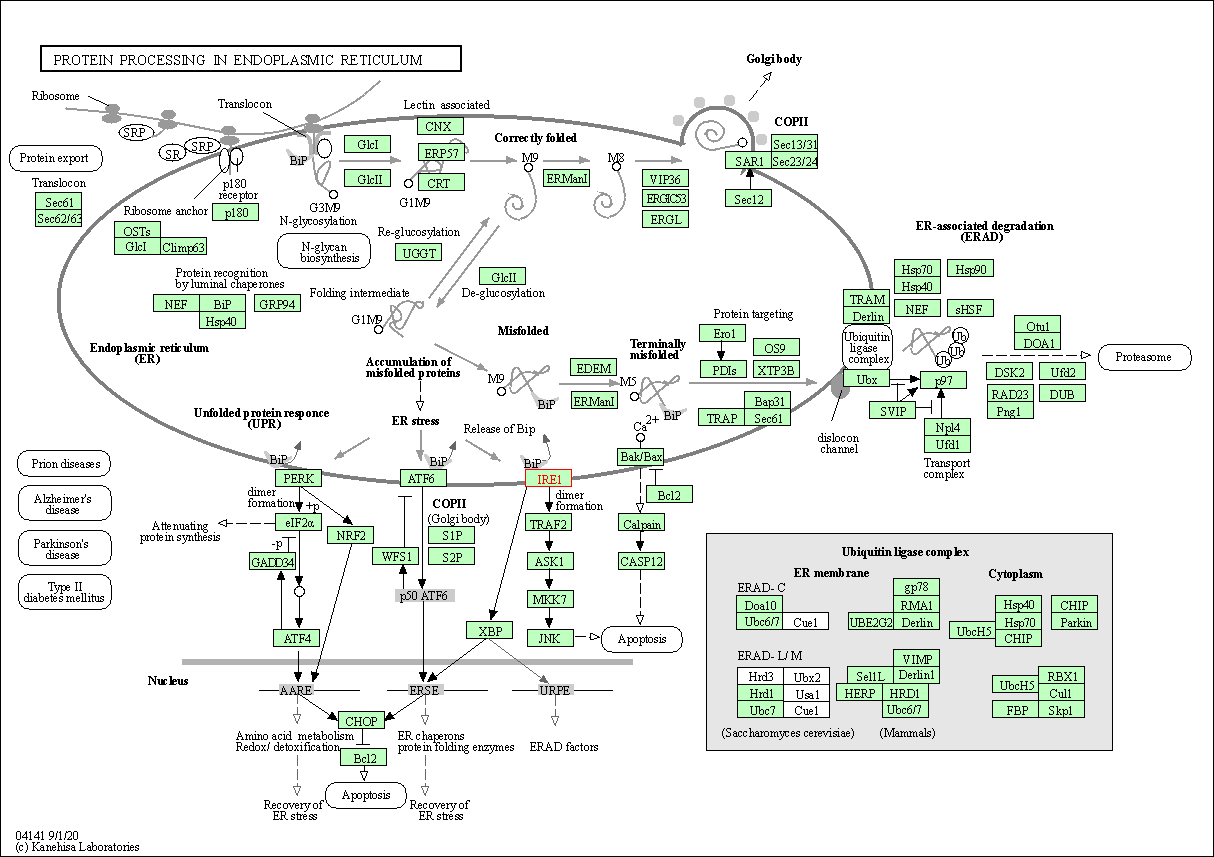
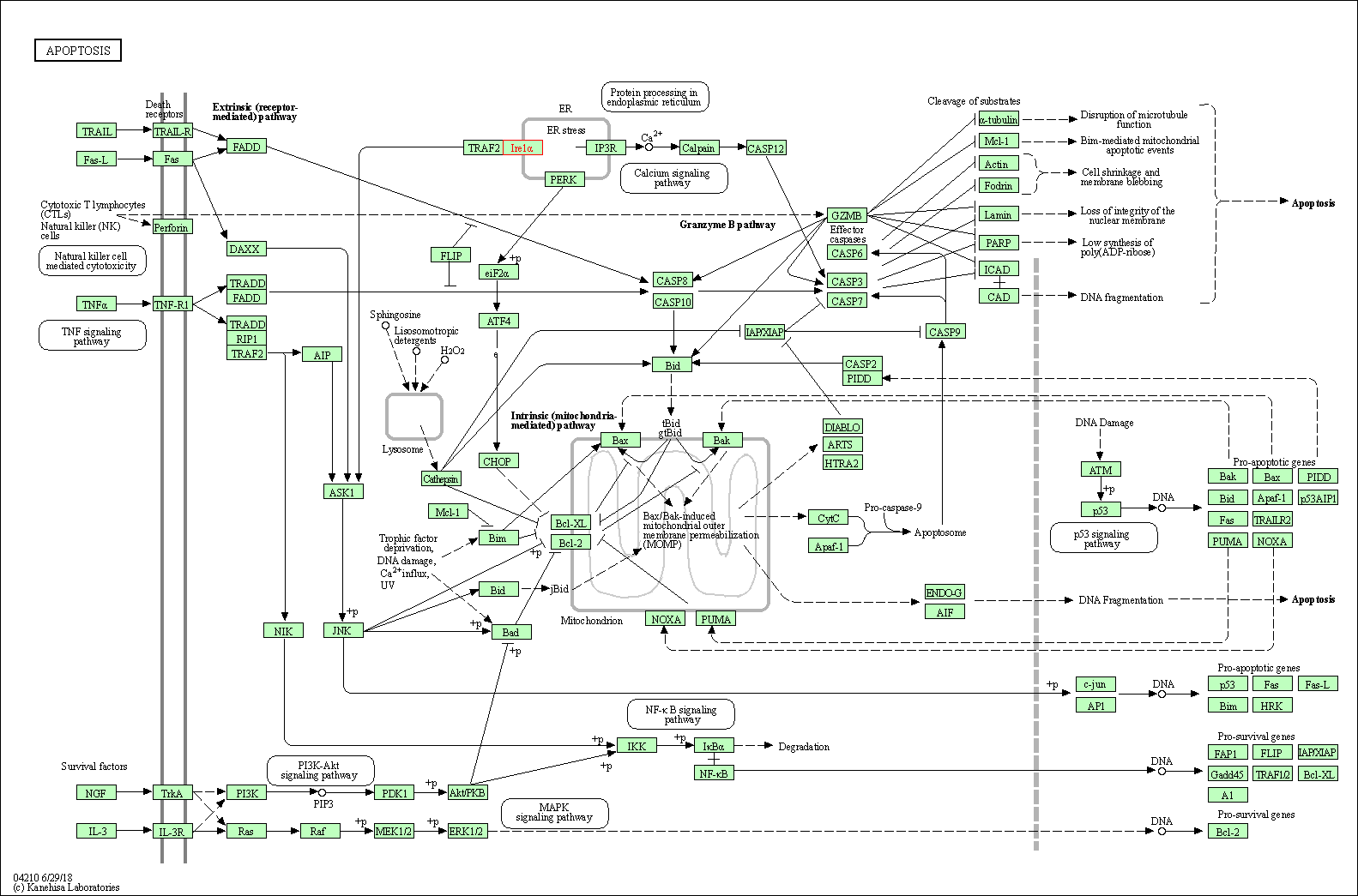
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Autophagy - animal | hsa04140 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |

|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Degree | 9 | Degree centrality | 9.67E-04 | Betweenness centrality | 4.59E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.29E-01 | Radiality | 1.40E+01 | Clustering coefficient | 1.39E-01 |
| Neighborhood connectivity | 4.48E+01 | Topological coefficient | 1.30E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Synthesis of novel tricyclic chromenone-based inhibitors of IRE-1 RNase activity. J Med Chem. 2014 May 22;57(10):4289-301. | |||||
| REF 2 | ClinicalTrials.gov (NCT03950570) ORIN1001 in Patients With Advanced Solid Tumors and Relapsed Refractory Metastatic Breast Cancer. U.S. National Institutes of Health. | |||||
| REF 3 | Clinical pipeline report, company report or official report of Orinove. | |||||
| REF 4 | IRE-1 inhibitors. US9040714. | |||||
| REF 5 | IRE-1 inhibitors. US9693992. | |||||
| REF 6 | IRE-1 inhibitors. US8614253. | |||||
| REF 7 | IRE-1 inhibitors. US9241942. | |||||
| REF 8 | IRE-1 inhibitors. US9493435. | |||||
| REF 9 | Quercetin ameliorates HFD-induced NAFLD by promoting hepatic VLDL assembly and lipophagy via the IRE1a/XBP1s pathway. Food Chem Toxicol. 2018 Apr;114:52-60. | |||||
| REF 10 | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1Alpha Inhibitory Activity. ACS Med Chem Lett. 2020 Oct 16;11(12):2389-2396. | |||||
| REF 11 | Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011 Mar 2;30(5):894-905. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

